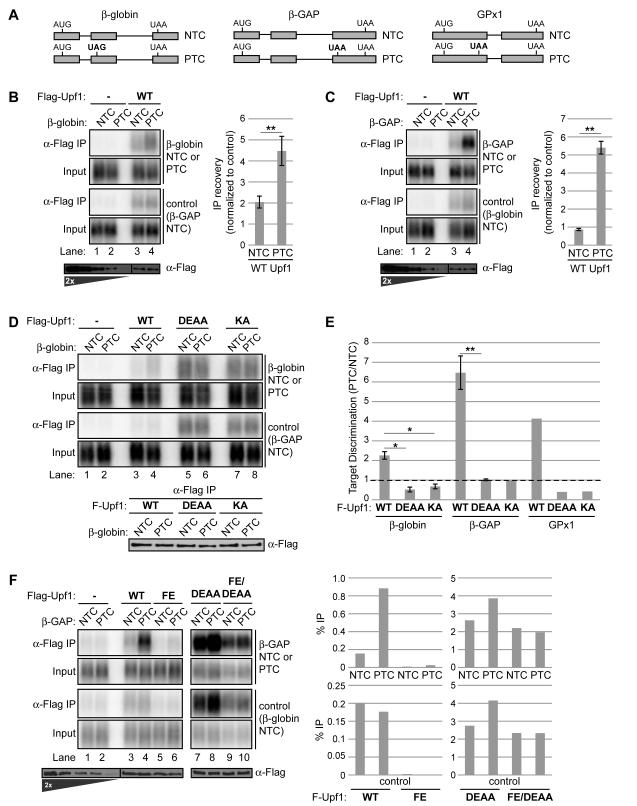
Figure 1. Upf1 ATPase cycle mutants are defective in selective NMD target association.
(A) NMD target (PTC) and non-target (NTC) mRNA reporter pairs used in RNA-IP assays based on human β-globin, β-globin with an insertion from GAPDH (β-GAP), or GPx1. The NMD-inducing termination codon is denoted in bold.
(B) Northerns of β-globin and control mRNAs in input (0.5%) samples or coprecipitated with Flag-Upf1 using anti-Flag antibody (α-Flag IP). Flag-Upf1 recovered in IPs is shown alongside a two-fold titration of input on anti-Flag Westerns below Northerns. The graph on the right represents recovery of β-globin mRNAs (NTC or PTC) with Flag-Upf1 over input normalized to recovery of the internal control (lanes 3,4), after subtraction of background in negative control IPs (lanes 1,2). Data are represented as mean +/− SEM for five biological replicates.
(C) Similar to panel B for RNA-IPs of β-GAP mRNAs.
(D) Similar to panel B, comparing IPs with Flag-Upf1 WT, D637A/E638A (DEAA), or K498A (KA). Western of recovered Flag-Upf1 variants is shown below Northerns.
(E) Graph representing the ratio of normalized IP recovery for NMD target (PTC) to non-target (NTC) mRNAs with the indicated Upf1 variants. A value of one, denoted by the dotted line, reflects an absence of discrimination between target and non-target. Data are represented as mean +/− SEM for three to four biological repeats.
(F) Similar to panel C but without normalization, comparing percent IP recovery of β-GAP mRNAs and the internal control by Flag-Upf1 WT, F192E (FE), DEAA or DEAA/FE mutants.
Asterisks denote P-values: *≤0.05, **≤0.001 (paired student’s t-test, two-tailed).
See also Figure S1