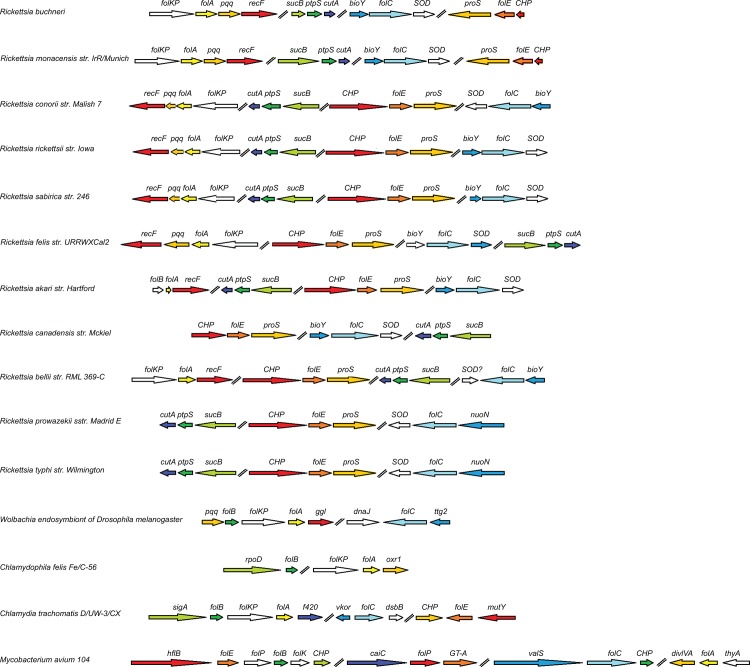
Fig 4. Physical clustering of folate biosynthetic genes across Rickettsia genus and related bacterial species.
ORFs and coding strand are indicated by arrows. The lengths of the arrows represent the lengths of the genes, with a scale of 0.1 inch = 100 bp. Adjacent arrows represent proximal loci, while the double lines between some arrows represent non-adjacency between loci. The names above the arrows represent the gene name; CHP gene encodes for conserved hypothetical protein with no known function; Abbreviations not described in the text: proS, prolyl-tRNA synthetase; sod, superoxide dismutase; bioY, biotin synthesis BioY protein; cutA, divalent cation tolerance protein; sucB, dihydrolipoamide succinyltransferase; nuoN, NADH dehydrogenase subunit N; ggl, gamma-glutamate ligase; dnaJ, molecular chaperone DnaJ; ttg2, toluene tolerance protein; rpoD, RNA polymerase sigma factor; oxr1, oxidoreductase 1; thyA, thymidylate synthetase; mutY, adenine DNA glycosylase; dsbB, disulfide bond formation protein; vkor, vitamin K epoxide; f420, coenzyme f420 hydrogenase; sigA, RNA polymerase sigma factor RpoD; hflB, ATP-dependent metalloprotease; caiC, crotonobetaine/carnitine-CoA ligase; GT-A, beta-1,4-N-acetyl-galactosaminyl transferase 1; valS, valyl-tRNA synthetase; divlVA, cell division protein DivlVA.