Abstract
Understanding the mechanisms producing low dose ionizing radiation specific biological effects represents one of the major challenges of radiation biology. Although experimental evidence does suggest that various molecular stress response pathways may be involved in the production of low dose effects, much of the detail of those mechanisms remains elusive. We hypothesized that the regulation of various stress response pathways upon irradiation may differ from one another in complex dose-response manners, causing the specific and subtle low dose radiation effects. In the present study, the transcription level of 22 genes involved in stress responses were analyzed using RT-qPCR in normal human fibroblasts exposed to a range of gamma-doses from 1 to 200 cGy. Using the alkali comet assay, we also measured the level of DNA damages in dose-response and time-course experiments. We found non-linear dose responses for the repair of DNA damage after exposure to gamma-radiation. Alterations in gene expression were also not linear with dose for several of the genes examined and did not follow a single pattern. Rather, several patterns could be seen. Our results suggest a complex interplay of various stress response pathways triggered by low radiation doses, with various low dose thresholds for different genes.
Keywords: low doses, ionizing radiation, dose response, human fibroblasts, DNA repair, gene expression
INTRODUCTION
The biological effects of low doses of ionizing radiation (LDR) and chemical toxins have been a focus of research in radiation biology and toxicology for a few decades. Their importance and relevance is hard to underestimate since all life forms, including human, are constantly exposed to low level stress factors in their day to day life. Ever increasing medicinal use of ionizing radiation, such as computed tomography, and other additional irradiation events, like occupational and accidental exposures in nuclear industry, have all either significantly or potentially contributed to increase human exposure to LDR. The stochastic nature and the lack of reliable methodology for the detection of very subtle changes in biological systems expected after LDR exposures have been a serious obstacle to gaining a complete understanding of the LDR effects.
It is widely accepted that LDR triggers very specific biological responses that deviate from linearity typically seen after exposure to high radiation doses (HDR). Although, these phenomena, and suggested underlying mechanisms, have been excellently covered in a number of reviews (Bonner 2004; Matsumoto et al. 2007; Marples and Collis 2008; Dauer et al. 2010; Morgan and Bair 2013), there is a lack of consensus about the factors that define molecular, cellular, tissue and organism responses specifically to LDR. In general terms, a model suggested by Feinendegen and Pollycove (Pollycove and Feinendegen 2003; Feinendegen and Neumann 2005) and consisting of three major components that define the shape of a dose-response curve for the formation of DNA mutations and for the risk of cancer seems the most appropriate. According to the model, it is the sum of 1) a linear increase with radiation dose of the number of initial DNA damage sites, 2) a relatively high rate of endogenous DNA lesions due to oxygen metabolism and nutrient deficiencies, and 3) non-linear responses of cellular and tissue defence systems that results in a lack of linearity for mutations and cancer risks. The first two components of the hypothesized model are almost axioms and have been experimentally validated in numerous studies, whereas the third one is the subject of a continuous debate and controversy.
LDR specific gene expression alterations have been found to be different from those observed after high doses (Ding et al. 2005; Franco et al. 2005; Mezentsev and Amundson 2011), strongly suggesting that there are qualitative and quantitative differences in the way cells respond to LDR compared to HDR. The number of genes affected in human lymphocytes by LDR vs. HDR, in the range 5 to 500 mGy, was the same; however, the genes altered in both cases were different (Nosel et al. 2013). Differential responses to LDR, compared to HDR, were confirmed in a mouse in vivo study using various lymphocyte subpopulations (Bogdandi et al. 2010). Consistent with the hypothesis of fundamentally different cellular responses to LDR vs. HDR are results showing two different mechanisms for the G2 cell cycle arrest, one being executed by ATM-dependent signalling very early after irradiation with doses ≥ 0.3 Gy (Xu et al. 2002; Asaithamby and Chen 2009). It was suggested that the lack of the early ATM-dependent G2-phase checkpoint caused low dose hyper-radiosensitivity (Marples et al. 2003; Asaithamby and Chen 2009).
We believe that to further advance our understanding of the mechanisms of LDR specific effects, it is necessary to gain a detailed picture of how various cellular and molecular stress response pathways are altered by radiation in a dose dependent manner. Moreover, a detailed dose response with small dose increment seems to be one of the prerequisites for the success in this endeavour since low dose thresholds for various endpoints have been reported (Day et al. 2006; Mitchel et al. 2008; Neumaier et al. 2012). With this in mind, we examined dose responses of various cellular stress response pathways in normal human fibroblasts using the expression of a set of genes and DNA single strand breaks and alkali-labile sites and their repair as end points.
MATERIALS AND METHODS
Cells and irradiations
Normal human embryonic lung fibroblasts (HELF-104; derived from an 8-week embryo) were purchased from BioloT, St-Petersburg, Russia. The cells were maintained by weekly sub-culturing in Dulbecco’s modified Eagle’s medium (DMEM; HyClone, Thermo Scientific) supplemented with 10% fetal bovine serum (HyClone, Thermo Scientific) without antibiotics at 37°C in a 5% CO2 and 95% air atmosphere. All experiments were carried out on asynchronous log-phase cell cultures of passage 22-24. The fraction of senescence-associated β-galactosidase positive cells was assessed using the Senescence Cells Histochemical Staining Kit (Sigma-Aldrich, USA) and was 3.9±0.3% (500 cells scored from each of 3 flasks). Subconfluent (50-70%) human lung fibroblasts were exposed to gamma-radiation doses of 0 (sham-irradiation), 1, 3, 5, 9, 12, 15, 20, 50, 100 and 200 cGy at 20°C using a «Rokus-AM» (Granit-Electron, Russia) device equipped with a 60Co source at a dose rate of 1 cGy/sec. Dose rate was measured and controlled using a DOSE-1 electrometer device equipped with an ionization chamber detector (IBA Dosimetry, Germany). Cells to be fixed at 5 min and 4 h after irradiation were exposed in normal growth DMEM and removed back to a CO2-incubator. For cells to be fixed immediately after irradiation (t = 1 min for the comet assay), DMEM was first substituted with ice-cooled phosphate buffered saline (PBS; pH 7.4, 10 mM phosphate buffer, 2.7 mM KCl, 137 mM NaCl) followed by the irradiation. Immediately before fixation, fibroblast monolayers were rinsed once with ice-cooled PBS and any residual liquid was removed from flasks. The fixation process consisted of snap-freezing by immersing the culture flasks into liquid nitrogen. The flasks were subsequently removed and stored at -85°C until analysis.
To measure initial levels of radiation-induced DNA damage, cells were immobilized into low melting point agarose on slides as described below in “The comet assay” sub-section and irradiated using the doses and dose rates identical to those used for irradiating cell cultures in flasks.
Real time PCR
Cells frozen at 4 h after irradiation with various doses were thawed on ice and RNA was extracted using Aurum Total RNA Mini Kit (BioRad, Hercules, CA, USA) as per manufacturer’s instructions. Extracted RNA was immediately quantified and its quality evaluated using a lab-on-chip capillary automated electrophoresis system Experion (Bio-Rad). Samples with an RNA Quality Index of 9 and more were used for further experimentation. One microgram of total RNA per sample was reverse transcribed into first strand cDNA using Maxima First Strand cDNA Synthesis Kit (Thermo Scientific, Rockford, IL, USA) as per manufacturer’s recommendations. All real time amplification reactions were conducted on a CFX96 PCR Detection System (Bio-Rad). The reaction mix contained 20 ng first strand cDNA, primers (300 nM each) and Maxima SYBR Green qPCR Master Mix (Thermo Scientific) in a final volume of 20 μL. The following PCR cycling conditions were used: 95°C for 10 min, 40 cycles of 95°C for 15 sec, 60°C for 60 sec. Each reaction was carried out in two technical replicates and mean Ct values were used for the evaluation of the gene expression levels by comparing the results produced for the irradiated groups with those produced for the non-irradiated control. Relative expression was calculated using the ΔΔCt method by normalizing to the house keeping genes ACTB and GAPDH. Data were analyzed using CFX Manager (Bio-Rad) and Excel (Microsoft) software. Our study of gene expression changes after irradiation consisted of 7 independent experiments separated from each other by at least 2 weeks.
Sequences of all primers used in this study were taken from literature sources and are presented in Table 1. Primer specificity and the size of PCR products were validated by examining melting curves and migration of bands in agarose electrophoresis with ethidium bromide.
TABLE 1.
Summary of genes analyzed in this study, qPCR primer sequences and references for primers and for their modulation in response to irradiation.
| Name | Function | Primers | Primers ref. | Induction to irradiation ref. |
|---|---|---|---|---|
| DDB2 | DNA-damage recognition and repair | F: CCAACCAGTTTTACGCCTCCTC R: TGTCTCCTGTGACCACCATTCG |
Roy et al. 2010 | Amundson et al. 2000 |
| XPC | F: TCTTCGGAGGGCGATGAAAC R: ATGATGGACAGGCCAATAGC |
Renaud et al. 2011 | Amundson et al. 2000 | |
| RAD52 | F: AGTTTTGGGAATGCACTTGG R: TCGGCAGCTGTTGTATCTTG |
Ghosh and Krishna 2012 | Riches et al. 2010 | |
| APEX1 | F: AGCCTTTCGCAAGTTCCTGA R: GCGTGAAGCCAGCATTCTTT |
Obtulowicz et al. 2010 | Ramana et al. 1998 | |
| GSR | Antioxidant protection | F: ATCCCCGGTGCCAGCTTAGG R: AGCAATGTAACCTGCACCAACAA |
Corrales et al. 2011 | Kojima et al. 1998 |
| TXN | F: CTTTGGATCCATTTCCATC R: GCATTAATGTTTTATTGTCACG |
Kim et al. 2008 | Kojima et al. 1998 | |
| CDKN1A | Cell-cycle control | F: CAGCAGAGGAAGACCATGTG R: GGCGTTTGGAGTGGTAGAAA |
Ghosh and Krishna 2012 | Kis et al. 2006 |
| IER5 | F: CCGGGAACGTGGCTAACC R: TTCCGTAGGAGTCCCGAGAA |
Ding et al. 2009 |
Kis et al. 2006; Ding et al. 2009 |
|
| PCNA | F: TTGCACTGAGGTACCTGAACTT R: CCTTCTTCATCCTCGATCTTG |
Gaube et al. 2007 |
Heinloth et al. 2003; Kis et al. 2006 |
|
| CCNG1 | F: GTCCCATTGGCAACTGACTT R: TGACATGCCTTCAGTTGAGC |
Ye et al. 2012 | Heinloth et al. 2003 | |
| CCNG2 | F: CCCAGAACCTCCACAACAG R: GGTGCACTCTTGATCACTGG |
Gaube et al. 2007 | * | |
| CCNE1 | F: GAAATGGCCAAAATCGACAG R: TCTTTGTCAGGTGTGGGGA |
Etemadmoghadam et al. 2010 | * | |
| GADD45α | F: TGCGAGAACGACATCAACAT R: TCCCGGCAAAAACAAATAAG |
Ghosh and Krishna 2012 |
Heinloth et al. 2003; Kis et al. 2006 |
|
| BBC3 | Apoptosis | F: CTGTGAATCCTGTGCTCTGC R: TCCTCCCTCTTCCGAGATTT |
Sharma et al. 2009 | Kis et al. 2006 |
| TNFSF10 | F: GCTGAAGCAGATGCAGGACAAG R: CTGACGGAGTTGCCACTTGAC |
Cheng et al. 2007 | Gong and Almasan 2000 | |
| BAX | F: AGAGGATGATTGCCGCCGT R: CAACCACCCTGGTCTTGGAT |
Sharma et al. 2009 | Amundson et al. 1999a | |
| MDM2 | Multiple function in the stress response | F: CTGGCTCTGTGTGTAATAAGGGAG R: CCTGATCCAACCAATCACCTG |
Jain et al. 2012 |
Amundson et al. 1999b; Ding et al. 2005 |
| EGR1 | F: TAGGCGGCGATTTTTGTATG R: TATCCCATGGGCAATAAAGC |
Kerman et al. 2012 | Ding et al. 2005 | |
| DUSP1 | F: GCCACCATCTGCCTTGCTTAC R: CTCGGTCGAGCACAGCCATG |
Baniwal et al. 2010 | Ding et al. 2005 | |
| ATF3 | F: AGAAGGAACATTGCAGAGCTAAG R: GGATTCTAGAGGTACACAGGAAG |
Saha et al. 2012 | Amundson et al. 1999a | |
| c-FOS | F: CGAGCCCTTTGATGACTTCCT R: GGAGCGGGCTGTCTCAGA |
Saha et al. 2012 | Prasad et al. 1995 | |
| PBP74 | F: TCTGGACTGAATGTGCTTCG R: ATCCCCATTTGTGGATTTCA |
Bian et al. 2008 | Sadekova et al. 1997 | |
| GAPDH | Reference | F: ACACCCACTCCTCCACCTTTG R: GCTGTAGCCAAATTCGTTGTCATAC |
Cheng et al. 2007 | |
| ACTB | F: GCGCGGCTACAGCTTCA R: CTTAATGTCACGCACGATTTCC |
Ding et al. 2009 |
*, no data for the gene expression modulation by radiation; added to the list due to their involvement in the cell cycle regulation and a cyclic expression pattern during progression through cell cycle phases (Lew et al. 1991; Horne et al. 1996).
The comet assay
DNA damage was evaluated using the alkaline comet assay that detects DNA single strand breaks, double strand breaks and alkali-labile sites (Tice et al. 2000). Cells in flasks were removed from -85°C storage and thawed immediately by a hot air stream directed to the bottom of a flask containing a cell monolayer. Preliminary experiments confirmed that the freezing and thawing of cells as described in this study did not introduce additional DNA damage (see Results for detail). Three hundred microlitres of cold PBS supplemented with 5 mM EDTA was immediately added to cells. The cells were detached from the plastic surface by a rubber cell scraper and the resulting cell suspension was mixed with 700 μL of 1.1% low melting point agarose (Sigma-Aldrich) prepared in PBS at pH 7.4 and 37.5°C. Then 100 μL of the resulting fibroblasts-agarose mix was placed on top of a glass microscope slide pre-coated with 1% normal melting point agarose (Sigma-Aldrich) and mounted with a cover-slip. Followed by a 5 min incubation at 4°C, cover-slips were gently removed and slides were immersed into a lysis buffer (2.5 M NaCl, 100 mM Na2EDTA, 10 mM Tris-HCl, pH 10.0, 10% DMSO, 1% Triton X-100) and incubated at 4°C overnight. All manipulations with cells during the initial steps of the process and up to an overnight lysis were done very quickly. However, to evaluate a possible contribution of the initial steps on the measurable levels of DNA damage (possible DNA repair), a dose response for the initial DNA damage levels was measured in agarose-embedded cells. To this end, the fibroblasts were trypsinized and rinsed twice in culture medium using centrifugation. After the second rinse, cells were re-suspended at a concentration 2×104 cells/mL and 300 μL of cell suspension were mixed with 700 μL of 1.1% low melting point agarose prepared in PBS and kept at 37oC. One hundred microlitters of the resulting cell-agarose mix was dispensed on top of a microscope slide pre-coated with 1% normal melting point agarose and mounted with a cover-slip. The slides were then incubated at 4°C for agarose solidification, placed inside petri dishes pre-filled with cold culture medium, ensuring that the slides are covered by at least 5 mm of medium, and brought to the irradiation room. During the irradiation, the samples were kept on ice to minimize DNA repair. Immediately after the irradiation (within 30 sec), the slides were immersed into a cold lysis buffer for an overnight incubation at 4°C. Starting from this point, slides prepared by both methods (irradiation of cell monolayers vs. irradiation of cells embedded into agarose) were processed identically. They were then incubated in the alkaline electrophoresis buffer (300 mM NaOH, 1 mM EDTA, pH>13) for 40 min at 4°C for DNA unwinding. Next, the slides were subjected to electrophoresis at 1 V/cm, 300 mA at 4°C for 25 min in fresh alkaline electrophoresis buffer. Following an extensive rinse in the neutralizing buffer (0.1 M Tris-HCl, pH 7.5) and then in bi-distilled water for 15 min, both at 4°C, the cells were fixed in ethanol for 10 min. The slides were then dried and 100 μL of 2 μg/mL ethidium bromide solution was added to the slides (Sigma-Aldrich). Cover slips were mounted on the slides and sealed for imaging with nail polish. The resulting comets were visualized using a fluorescence microscope Axioscop-40 FL (Carl Zeiss, Jena, Germany), at 200X magnification. Images were captured using a CCD camera AxioCam ICm 1 and an AxioVision software package (Carl Zeiss) at a 1338 x 1038 pixels resolution. Olive tail moment (OTM) that captures both the smallest detectable size of migrating DNA and the number of DNA breaks (Marples and Collis 2008) was calculated using the CometScore Pro software (TriTek Corp, USA). The mean value of OTM from 100 comets per slide was calculated and used as an index of DNA damage. Experiments with irradiating cell monolayers were carried out in 4 replicates, each separated by at least 2 weeks from one another. Each biological replicate consisted of 3 slides per dose. The experiment with irradiating cells embedded into agarose was done once and contained 9 slides per dose.
Statistical analyses
The gene expression experiments were carried out in 7 independent replicates. Within each independent replicate, mean Ct value of two technical replicates were used to calculate normalized relative expression in irradiated groups compared to the control one using the ΔΔCt method. Gene expression data were tested by the Grubb’s method for the presence of outliers and the detected outliers were removed from subsequent analyses. The resulting data set consisted of 7 independent replicates for most genes and 6 independent replicates for a few genes and irradiation groups. Due to normalization of the data to control values, parametric statistics could not be applied. Instead, for pairwise comparisons of each dose to its non-irradiated control, we used the Wilcoxon signed-rank test. The minimum possible p value for two dependent samples with N=7 is 0.018. This test also requires the Bonferoni correction or the False Discovery Rate correction. It follows then that, given the design of the study (10 pairwise comparisons with the control resulting in the minimum p value of 0.005), statistical significance is not achievable regardless of the difference between treatment and control groups. Therefore, in the figures we presented the probabilities of the null-hypothesis for every individual comparison. If no simple regression fit could be used for the data with statistical significance, only one dose in the entire range was allowed to produce a “false discovery”. Data were plotted using Excel (Microsoft) and Statistica v.6 (Statsoft) software packages.
The comet assay experiments were independently repeated 4 times. Three slides were prepared and analyzed per experimental replicate. Mean values of OTM from 100 comets per slide were calculated. Final mean values presented on the plots were, therefore, calculated from 12 slides obtained in 4 independent experiments. For a supplementary optimization experiment, when the cells were irradiated on slides embedded into agarose, mean OTM values were calculated from 9 slides per treatment within a single experiment. Standard deviations were used to evaluate errors. The levels of DNA damage obtained for 4 h or 5 min after irradiation were compared with the DNA damage level at 1 min using the Student t-test and Mann-Whitney U-test. Dose-response trends for DNA damage levels were analyzed using one-way ANOVA test separately for 1, 5 min and 4 h data sets. Statistical differences in DNA damage levels between individual dose points were evaluated using a multiple post-hoc test (Newman-Keuls).
Regression analysis was carried out using SPSS 15 software (SPSS, IBM). The statistical significance of the R2 coefficient increase due to the use of a linear fit compared to a polynomial fit was evaluated using the Hayek criterion.
RESULTS
Dose responses for the expression of stress response genes
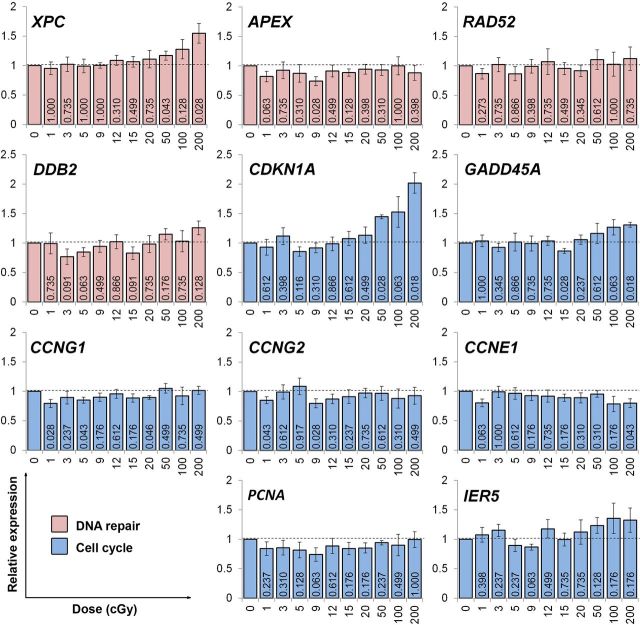
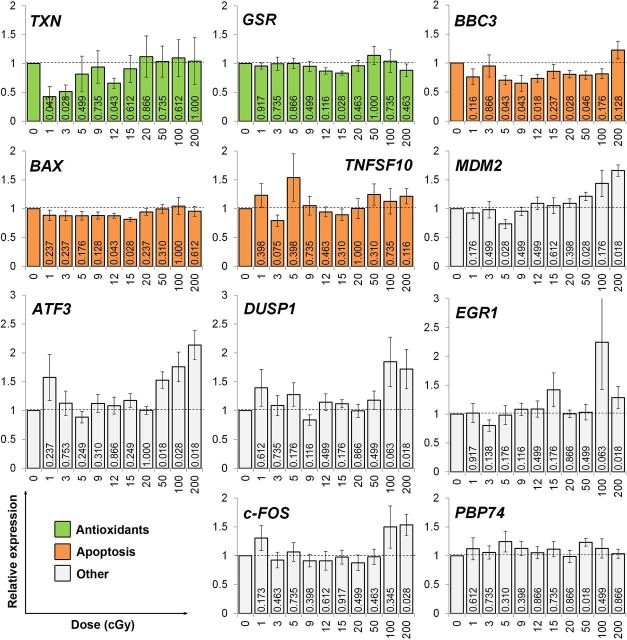
To examine molecular stress responses after irradiation with 1, 3, 5, 9, 12, 15, 20, 50, 100 and 200 cGy, we measured, using RT-qPCR, changes in the expression of a panel of 22 genes involved in various stress response functions. Only genes that have been experimentally shown to be transcriptionally modulated by radiation were chosen (references provided in Table 1). The obtained transcriptional dose responses at 4 h after irradiation are presented in Fig. 1 and Fig. 2 relative to the non-irradiated control. To increase statistical significance of the expression data, the experiments were carried out as 7 independent replicates. A few single data points were identified as outliers using the Grubb’s test and were excluded from the analyses.
FIG. 1.
Radiation dose responses for the expression of DNA repair and cell cycle genes. Human fibroblast cultures were exposed to the indicated doses of gamma-radiation, followed by 4 h incubation under normal growth conditions to allow cells to develop a transcriptional response. RNA was then extracted, reverse transcribed and the expression of the target genes was measured by real time PCR. Relative expression was calculated using the ΔCt method. A value of 2-ΔΔCt > 1 indicates an increase in gene expression compared to the non-irradiated control, whereas a value of 2-ΔΔCt < 1 indicates a decrease in gene expression compared to the non-irradiated control. Results of 7 independent experiments ± SD are shown. Numbers on the bars are probabilities of the null hypothesis calculated using the Wilcoxon matched pairs test. The color coding was used to identify stress response pathways/functions the genes have roles in: pink, DNA repair; blue, cell cycle.
FIG. 2.
Radiation dose responses for the expression of antioxidant, apoptosis and other stress response genes. Human fibroblast cultures were exposed to the indicated doses of gamma-radiation, allowed to develop transcriptional responses for 4 h and RNA was extracted and the expression of genes indicated was measured and quantified as described in Fig.1. Results of 7 independent experiments ± SD are shown. Numbers on the bars are probabilities of the null hypothesis calculated using the Wilcoxon matched pairs test. The color coding was used to identify stress response pathways/functions the genes have roles in: green, antioxidant; orange, apoptosis; white, other pathways.
Firstly, a number of genes showed no alterations in the expression following any of the doses used. Those include DNA repair genes DDB2, RAD52 and APEX, cell cycle regulators IER5 and PCNA, antioxidant gene GSR, a pro-apoptotic cytokine TNFSF10, transcriptional regulators EGR1, DUSP1 and c-FOS and a heat-shock gene PBP74. At the same time, the expression of CCNE1 was decreased slightly with increasing irradiation dose (R2= 0.384, SR = 0.064, F = 5.615, p = 0.042). The expression of the remaining genes was altered by irradiation, with some genes showing complex dose responses described below.
Secondly, the expression of 5 genes was increased in response to doses of 50-200 cGy in all 7 or 6 experimental replicates. The activation of all 5 genes could be best fit with a linear function (ATF3: R2 = 0.75, SR = 0.204, F = 26.9, p = 0.001; CDKN1A: R2 = 0.92, SR = 0.105, F = 101.6, p < 0.001; and XPC: R2 = 0.97, SR = 0.034, F = 246.5, p < 0.001; MDM2: R2 = 0.86, SR = 0.102, F = 53.8, p < 0.001; GADD45A: R2 = 0.73, SR = 0.074, F = 24.1, p = 0.044). The validity of the linear fits were confirmed using the Hayek’s criterion.
Thirdly, and most interestingly, the expression of the remaining 5 genes was found to decrease after the exposure to at least two low radiation doses in all 7 or 6 biological replicates. However, the observed gene expression down-regulation induced by LDR could not be fit by a simple correlation with dose. Thus, the dose responses for modulation of the TXN, CCNG1, CCNG2, BAX and BBC3 genes had complex, non-monotonic patterns. The most pronounced change of > 2 fold after as low as 1 cGy was found for the TXN gene encoding the antioxidant protein thioredoxin (Fig. 2). The other genes responding to low radiation doses in such a manner were cell cycle regulators CCNG1 and CCNG2 (Fig. 1) and pro-apoptotic genes BAX and BBC3 (Fig. 2).
DNA damage dose response and repair kinetics
Typically, when assessing initial DNA damage by the comet assay in suspended cells, such as lymphocytes, live cells are irradiated embedded into low melting point agarose on a slide and lysed immediately after the irradiation (Neumaier et al. 2012). However, this protocol may not be ideal for adherent cells, such as human fibroblasts used in this study, due to the fact that trypsinization itself introduces stress which may affect the cells response to subsequent irradiation. Even more importantly, if a study involves repair kinetics, i.e. requires an incubation of cells for certain time periods after irradiation under normal growth conditions, irradiation for an initial t = 0 and subsequent time points should be carried out in an identical way. It follows then that for human fibroblasts, that are adherent cells, irradiation should be conducted on monolayers, rather than suspended cells. We, therefore, optimized a method for irradiating human fibroblast cultures and immediately preserving them by snap-freezing in liquid nitrogen for subsequent processing for the comet assay. Cells frozen in this way can be stored at -85°C for extended periods of time, allowing processing for the comet assay a large number of samples in a single run and thus eliminating inter-experimental variability. To confirm that the freeze-thaw cycle as described in “Materials and Methods” does not introduce additional DNA damage, we compared the OTM value obtained for cells that were subjected to the freeze-thaw cycle to that for cells that were prepared freshly without freezing. The two OTM values were 1.83 ± 0.23 and 1.97 ± 0.24 (3 slides × 3 replicates per group) for the freeze-thaw sample and the control, respectively, and were not statistically different at p ≤ 0.05 (Student t-test). This result confirms that our experimental approach is appropriate for the measurement of DNA single strand breaks and alkali-labile sites induced by radiation and the repair kinetics in human fibroblast cultures.
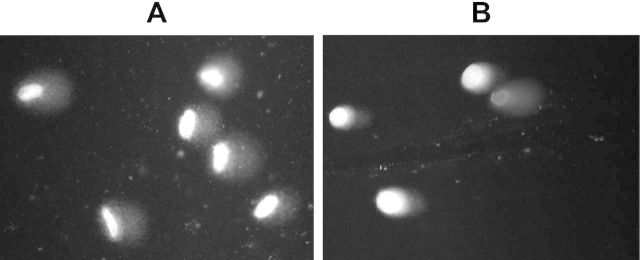
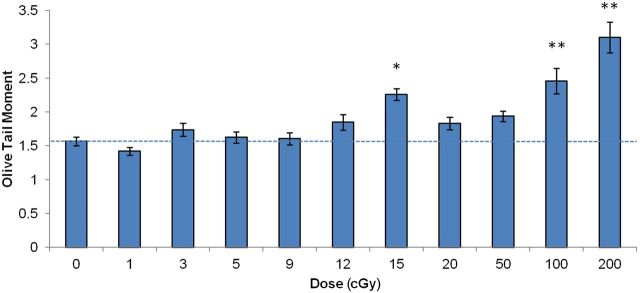
However, there were valid concerns that irradiating cell monolayers for the initial time point to evaluate the initial DNA damage level would still allow enough time before the lysis step to repair DNA damage. The minimum time period between the end of irradiation and a lysis was 1 min. This could potentially affect the sensitivity of our modification of the comet assay. Particularly prone to a bias introduced by this potential loss of sensitivity would be DNA repair kinetics data. To address those concerns, we conducted a dose-response experiment in which we irradiated agarose-embedded cells on ice to evaluate the initial DNA damage level. The first evident difference between the samples prepared from irradiated monolayers vs. irradiated agarose-embedded cells was the shape of the cells: higher heterogeneity and irregularity in shapes was found for the irradiated monolayers compared to cells irradiated in agarose (Fig. 3). Next, we compared dose-response relationships between the two methods. As expected, under the conditions when cells do not have time to repair damage (agarose-embedded cells), its production was linearly dependent on dose (R2 = 0.85, SR = 0.2, F = 51.9, p < 0.001). One-way ANOVA test confirmed the dose dependence of the OTM value (p < 0.001). Post-hoc analysis showed that significant changes in the DNA damage level were induced by doses of 15, 100 and 200 cGy (Fig. 4). With irradiated monolayers, overall DNA damaging effect of irradiation was significant (One-way ANOVA: p < 0.05 with post-hoc, p < 0.05 for 200 cGy; Fig. 5A, 1 min dataset). Regression analysis of the entire dose range produced a statistically significant linear fit with R2 = 0.77 (SR = 0.16, F = 33.7, p < 0.001). This result indicates that the experimental approach with irradiating monolayers has slightly decreased sensitivity compared to irradiating cells in agarose, yet a statistically significant linear dose response was observed for both approaches. Both slightly longer repair time and the shape of the cells could potentially contribute to such an effect. However, we reasoned that the necessity of preserving the optimal physiological conditions during repair experiments outweighed the observed loss in the sensitivity.
FIG. 3.
Representative images of DNA comets obtained from 200 cGy irradiated normal human fibroblasts. (A) Irradiated monolayers: HELF-104 cells were harvested using a cell scraper 1 min after the irradiation and processed as described in Materials and Methods. (B) Irradiated agarose-embedded cells: HELF-104 cells were trypsinized, resuspended and immobilized in low melting point agarose on microscope slides. Cells were then irradiated, immediately lysed and processed for the comet assay as described in Materials and Methods.
FIG. 4.
Radiation dose response for DNA damage in agarose-embedded normal human fibroblasts. Cells were irradiated as agarose-embedded suspensions after trypsinization, followed by an immediate lysis and processing for the comet assay as described in Materials and Methods. Mean values from 9 slides ± SD are shown. * and ** denote statistically significant effect of irradiation with p < 0.01 and p < 0.001, respectively (ANOVA with post-hoc analysis by Newman-Keuls test).
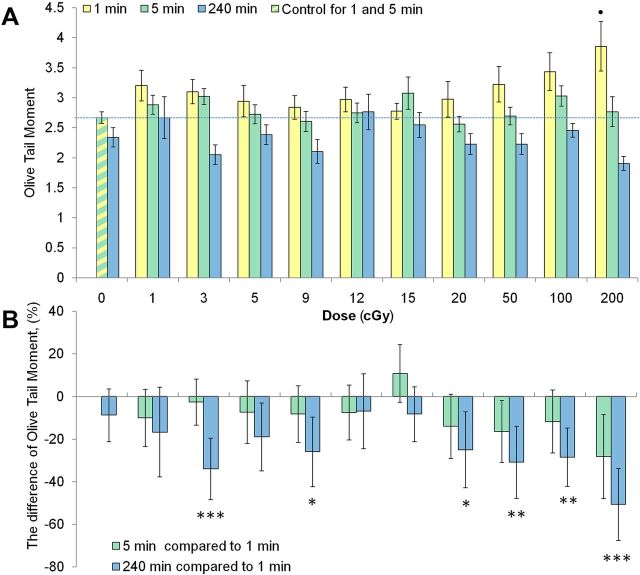
FIG. 5.
Radiation dose response for DNA damage and repair in monolayers of normal human fibroblasts. Cells were irradiated as monolayers under normal growth conditions. (A) Normal human fibroblast cultures were exposed to the indicated doses of gamma-radiation and fixed either immediately (1 min) or followed by an incubation under normal growth conditions for 5 min or 4 h to allow the repair of DNA damage. Cells were then processed for the alkaline comet assay and the OTM values were calculated and plotted. (B) To evaluate the extent of the induction of DNA repair, the residual level of DNA damage at 5 min and 4 h time points was compared to the initial level of DNA damage at 1 min. The plot shows the percentage difference for the OTM values between 5 min and 1 min datasets (4 min of repair time) and between 4 h and 1 min datasets (~4 h of repair time). Mean values from 4 independent experiments ± SD are shown. • denotes statistically significant effect with p < 0.05 (ANOVA with post-hoc analysis by Newman-Keuls test). *, **, and *** denote statistically significant difference between 4 h and 1 min datasets, with p < 0.05, < 0.01 and < 0.001, respectively (Student t-test).
Next series of experiments aimed at evaluating the repair potential of radiation-induced damage involved irradiating monolayer cell cultures and harvesting them for the comet assay at 1 min (the minimum technically possible time point as described above, which served as initial damage level), 5 min and 4 h post-irradiation. This experimental approach ensures that cells are allowed to repair DNA damage under normal physiological conditions as opposed to conditions where agarose immobilization during repair times may introduce additional stress and biased DNA repair responses.
DNA damage produced at 1 and 5 min after irradiation was compared to a combined control generated for 1 and 5 min time points (the green-yellow bar in Fig 5A), whereas the 4 h time points were compared to their own 4 h control. Those control samples were made of sham-irradiated cell cultures incubated in parallel with the irradiated ones for 4 h. No significant increases in DNA damage levels could be found at 5 min or 4 h after irradiation. This indicates fast repair of DNA single strand breaks and alkali-labile sites in normal human fibroblasts. Besides, this result provides further support for the DNA repair as the main cause of somewhat lower sensitivity of the comet assay seen when comparing dose responses produced for agarose-embedded cells (Fig. 4) and for monolayer cell cultures (Fig. 5A).
Using the 5 min and 4 h data, we evaluated the DNA damage repair kinetics by comparing the OTM values to those obtained at 1 min for each radiation dose. The difference was expressed as percentage difference and is shown in Fig. 5B. When the DNA damage levels were compared between 1 min and 4 h time points within each treatment group, statistically significant differences were found for 3, 9, 20, 50 and 200 cGy. It was not surprising to observe that DNA single strand breaks and alkali-labile sites induced by relatively high doses of 100 and 200 cGy were efficiently repaired by 4 h post-irradiation. However, the finding of active DNA repair after low doses of 3, 9 and 20 cGy, and a higher dose of 50 cGy was somewhat unexpected since the doses themselves produced low or no increase in the damage levels (Fig. 5A). Another striking observation was that two doses in the middle of the dose range examined, e.g. 12 and 15 cGy, did not induce any DNA repair, which could be seen by comparing their delta values to those of the control. These results suggest a complex, non-linear DNA damage inducible repair response to LDR.
DISCUSSION
Conventional wisdom dictates that the level of radiation-induced damage to macromolecules increases in a linear dose-dependent manner and experimental evidence of that for DNA damage has been obtained (Rothkamm and Löbrich 2003; Osipov et al. 2004). However, one of the basic features of the cell is its ability to respond to external stimuli, including damage inflicted by exposure to ionizing radiation. These cellular responses undoubtedly affect the ultimate end points typically assessed in radiobiological studies. Furthermore, the more separated in time and organizational level the end point from the initial event of molecular damage to DNA is, the less a linear dose-effect relationship is expected. Indeed, numerous reports confirmed the lack of linearity in radiation dose responses at low doses for such end points as micronuclei (Słonina et al. 2006), chromosomal aberrations (Nasonova et al. 2006), cellular clonogenic survival (Marples and Joiner 1993) and others. The present work examining a dose range of 1-200 cGy had a particular focus on low doses between 1 and 20 cGy. We should stipulate that a dose of 50 cGy is assigned in this discussion to a high dose range. Although this dose is not considered high by all authors, this is done for two main reasons, one being that the context of a potential significance of this work is in the radiological protection field. In this field, doses below 20 cGy are considered low and doses greater or equal to 50 cGy are considered high (UNSCEAR 2012). The second reason is to avoid unnecessary complication by introducing an intermediate dose range that would be represented only by this single dose of 50 cGy. Our present results demonstrated that early (4 h) changes in the expression of a selected panel of genes regulating major stress response pathways are different for LDR compared to HDR in that: a) non-overlapping groups of genes respond to LDR vs. HDR; b) mostly down-regulation for LDR, as opposed to up-regulation for HDR was observed; and c) complex, non-monotonic relationships for LDR, as opposed to linear ones for HDR were seen. Some of these observations are consistent with literature data showing that a modulation of transcriptional gene activity was qualitatively different after LDR compared to HDR exposures (Ding et al. 2005; Franco et al. 2005; Nosel et al. 2013). However, in majority of gene expression studies, two contrasting doses from low and high dose ranges were used to compare the resulting transcriptional changes. Obviously, this approach does not allow dose-response relationships to be evaluated, which, given the arbitrariness of the dose and cell type choice and possible variations in responsiveness within a low dose range itself, represents a serious limitation. In this study, we examined gene responses over a wide dose range with a relatively small dose increment between 1-200 cGy. Combined with a high number of independent experimental replicates, this allowed us to generate a detailed transcriptional dose response for a set of genes with confirmed roles in cellular stress response pathways. Thus, we found a linear correlation with dose for CDKN1A, GADD45A, ATF3, XPC and MDM2, with activation seen mostly at high doses. This result is consistent with the findings of other researchers showing linear dose responses for CDKN1A, GADD45A, ATF3 and MDM2 in human myeloid leukemia ML-1 cells (Amundson et al. 1999b), and for CDKN1A и XPC in human peripheral blood lymphocytes (Amundson et al. 2000). Combined with the published data, our results obtained in human fibroblasts indicate the versatility of the up-regulation of those genes in different human tissues in response to radiation.
Rather irregular, yet consistent in multiple replicates carried out in this study, were responses of CCNG1, CCNG2, BAX, BBC3 and TXN genes. In general, these genes were down-regulated by LDR and were not affected by HDR, explaining the lack of the reverse correlation with dose. Remarkably, different genes from this group had different low dose thresholds for down-regulation. Thus, CCNG1, CCNG2 and TXN were suppressed by as low as 1 cGy, whereas the response for BBC3 was reliably seen at 5 cGy and for BAX at 12 cGy. This result providing additional information within the low dose range itself highlights the value of having a small dose increment for dose-response studies. Furthermore, the genes responsive to LDR were not actually responsive to all low doses. Instead, rather complicated patterns could be seen. For example, CCNG1 was suppressed by 1, 5 and 20 cGy, but not by 3, 9, 12, 15 cGy. CCNG2 responded to 1 and 9 cGy, but not to 3, 5, 12, 15 or 20 cGy. We would agree that such, at a glance, random behaviours of data points may partially be explained by a high rate of false positives. Complicating the issue, small increment and resulting high number of pairwise comparisons that had to be made undoubtedly compromise the statistical power of our results. Indeed, statistics using non-parametric pairwise criteria is the only correct way of calculating significance of comparisons between individual doses and control groups. We calculated that statistical significance using this approach is not achievable at all, no matter how different the control and experimental groups are (e.g. 1000-fold difference would still be not significant!). Obviously, this is somewhat a counter-intuitive result that affects the interpretation of the biology behind our data. Given the consistency with which the described gene expression changes were observed in multiple (6-7) independent experiments separated in time, we argue that the revealed trends have biological significance. This is further supported by the analysis of functions of the genes found to be modulated by LDR in this study. Cell cycle, apoptosis and antioxidant protection have a significant impact on the systemic end points for which non-linear dose responses have been shown. Both CCNG1 and CCNG2 are involved in cell cycle regulation via various pathways (Zimmermann et al. 2012; Zhao et al. 2003; Ohtsuka et al. 2004). Obviously, one would not expect drastic changes in cell cycle phases distribution after as low as 1 cGy. Yet, rather weak changes in cell proliferation may well be expected, consistent with our unpublished observations of enhanced cell growth of LDR treated normal human cells relative to the non-irradiated control. Other authors have also showed that 10 cGy irradiation of a 3D cell culture mimicking human epidermis resulted in the down-regulation of cyclins A2, B1 and B2, confirming a potentially significant role of cell cycle regulation in the responses to LDR (Mezentsev and Amundson 2011). Slightly higher doses (5-15 cGy) modulated two apoptosis regulators BAX and BBC3, indicating that apoptosis pathway may play a role in systemic (e.g. survival) responses to LDR. Finally, the most pronounced alterations produced by LDR (up to 2-fold down-regulation) were found for the TXN gene encoding a tioredoxin protein involved in the cellular protection against oxidative stress and apoptosis (Li et al. 2013).
Unlike most studies examining transcriptional profiles in response to radiation, we conducted parallel experiments to measure DNA damage and repair rates. This allows an attempt to correlate molecular transcriptional responses with a physiological DNA damage end point. Although we found complex, non-monotonic dose responses for both end points, there was no clear correlation between the DNA damage and the gene expression responses.
Non-linearity in the DNA repair responses measured as a difference between the initial DNA damage levels at 1 min and the residual levels at 4 h was observed. Thus, we found an inducible repair response after low doses of 3 and 9 cGy and after higher doses of 20, 50, 100 and 200 cGy, but not after the doses of 12 and 15 cGy. There was also a statistically not significant trend towards inducible DNA repair after 1 and 5 cGy doses (Fig. 5B). Noteworthy, the described trends in the DNA repair responses were confirmed by both parametric (Student t-test) and non-parametric (Mann-Witney U-test) statistics. There is controversial evidence with respect to the cell’s ability to detect and repair DNA damage at LDR. It was hypothesized and experimental evidence obtained that LDR-induced DNA damage is poorly sensed and remains unrepaired for long periods of time (Rothkamm and Löbrich 2003; Grudzenski et al. 2010). Challenging that concept are studies demonstrating normal or even enhanced DNA repair response after exposure to LDR (Asaithamby and Chen 2009; Neumaier et al. 2012). Apparently, mechanisms similar to the ones driving DNA repair induction in this study were causing the suppression of the DNA double strand break levels below the background one reported for mice chronically exposed to very low dose rate gamma-irradiation (Osipov et al. 2013). It appears that apoptotic elimination of a highly damaged or hyper-sensitive sub-population of cells would not play a major role in the observed effect since fibroblasts do not readily undergo apoptosis after irradiation (Mirzayans et al. 2013). Besides, the 4-h time point used in our study would not provide enough time for a radiation-induced apoptosis. It follows then that this effect may be considered in terms of radiation adaptive response terminology. Noteworthy, not all the doses in the range between 0 and 50 cGy lead to the repair activation. Doses of 1, 5, 12 and 15 cGy did not produce statistically significant repair responses. These data strongly suggest non-linear, highly complex repair responses to LDR and are consistent with the hypothesis that it is the defence response component (the component #3 in the model suggested by Pollycove and Feinendegen (2003)) that causes non-linearity of mutation and cancer risks after exposure to LDR.
Apparently, it is the complexity and non-monotonic pattern of the molecular stress responses and the inducibility of DNA repair demonstrated in the present study that contributes most to non-linear responses typically seen for integral end points, such as gross chromosomal aberrations (Marples and Joiner 1993; Nasonova et al. 2006) or cell survival (Grudzenski et al. 2010). Suggesting a particular, single molecular mechanism that would produce such an outcome does not seem reasonable. Instead, a sophisticated dose dependent interplay of various cellular stress response systems should be involved.
Although this study does not provide a direct link between the observed LDR-induced molecular changes at the transcription level and delayed systemic effects found in other studies, indirectly our results point to such a link. Indeed, our selection of genes included the ones with confirmed roles in radiation responses and covered the most essential pathways in those responses. It is therefore expected that changes, however subtle, in the regulation of the genes after exposure to LDR would have an effects on the ultimate physiological or systemic outcome. Supporting this notion are findings that knock-down or knock-out of single genes may affect such integral end points as clonogenic survival after irradiation (Criswell et al. 2005), life span in control or radiation-exposed organisms (Moskalev et al. 2011). Moreover, modulation of the expression of a single gene in one tissue only was sufficient to increase the life span of D. melanogaster (Plyusnina et al. 2011; Shaposhnikov et al. 2011). Obviously, post-translational protein changes and some other molecular changes, not assessed in this study, play very important roles in cellular responses to radiation. Yet, they are often intricately linked to transcriptional changes, being most often upstream of the gene expression changes assessed in this study. In this respect, our gene expression data may, to some degree, reflect those post-translational responses to radiation that are not related to direct functions, such as DNA repair. Undoubtedly, it would be interesting to examine dose responses for global post-translational changes, e.g. phosphorylation. Yet, this may become a focus of future studies. Overall, it is advisable to consider our data as single representative slice of a multifaceted big picture that is much wider (number of genes) and deeper (various levels of regulation, such as post-translational modifications, chromatin modification, miRNA levels) in nature.
CONCLUSIONS
Complex modulation of the expression of 11 out of 22 stress response genes with confirmed roles in the radiation responses was observed, with LDR causing the suppression and HDR predominantly resulting in the induction of the selected genes. Noteworthy, the dose-response curves for gene transcription levels varied qualitatively for different genes. Additionally, new evidence for non-linear, highly complex repair responses to LDR was obtained.
Our results contribute towards an improved understanding of the biological effects of LDR and provide further evidence to suggest that the LNT model may not be the best model to predict the consequences of radiation-induced outcomes over a wide range of doses.
ACKNOWLEDGMENTS
We thank Alexander Y. Moseyko for help with dosimetry and irradiations, Nicholas Priest for reading the manuscript and helpful discussions and Tatiana A. Maystrenko for advice on statistical analysis of the data. Funding for this study was provided by the Russian Fund for Fundamental Research Grant 13-04-01750, Ural Branch of Russian Academy of Sciences Grant for Young Scientists 13-4-НП-180 and by Atomic Energy of Canada Limited.
Footnotes
AUTHOR CONTRIBUTIONS: Conceived and designed the experiments: I.O.V., D.Y.K. Performed the experiments: I.O.V., D.M.Sh., Y.I.P., E.S.B., O.A.Sh., A.V.E., A.V.K., O.V.E. Analyzed and interpreted the data: I.O.V., D.Y.K., E.S.B. Wrote the manuscript: I.O.V., D.Y.K.
CONFLICTS OF INTEREST: The authors declare no conflict of interest.
REFERENCES
- Amundson SA, Bittner M, Chen Y, Trent J, Meltzer P, Fornace AJ., Jr 1999a. Fluorescent cDNA microarray hybridization reveals complexity and heterogeneity of cellular genotoxic stress responses. Oncogene 18: 3666–3672. [DOI] [PubMed] [Google Scholar]
- Amundson SA, Do KT, Fornace AJ., Jr 1999b. Induction of stress genes by low doses of gamma rays. Radiat Res 152: 225–231. [PubMed] [Google Scholar]
- Amundson SA, Do KT, Shahab S, Bittner M, Meltzer P, Trent J, Fornace AJ., Jr 2000. Identification of potential mRNA biomarkers in peripheral blood lymphocytes for human exposure to ionizing radiation. Radiat Res 154: 342–346. [DOI] [PubMed] [Google Scholar]
- Asaithamby A, Chen DJ. 2009. Cellular responses to DNA double-strand breaks after low-dose gamma-irradiation. Nucleic Acids Res 37: 3912–3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baniwal SK, Khalid O, Gabet Y, Shah RR, Purcell DJ, Mav D, Kohn-Gabet AE, Shi Y, Coetzee GA, Frenkel B. 2010. Runx2 transcriptome of prostate cancer cells: insights into invasiveness and bone metastasis. Mol Cancer 9: 258–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian Q, Fernandes AF, Taylor A, Wu M, Pereira P, Shang F. 2008. Expression of K6W-ubiquitin in lens epithelial cells leads to upregulation of a broad spectrum of molecular chaperones. Mol Vis 14: 403–412. [PMC free article] [PubMed] [Google Scholar]
- Bogdandi EN, Balogh A, Felgyinszki N, Szatmari T, Persa E, Hildebrandt G, Sáfrány G, Lumniczky K. 2010. Effects of low-dose radiation on the immune system of mice after total-body irradiation. Radiat Res 174: 480–489. [DOI] [PubMed] [Google Scholar]
- Bonner WM. 2004. Phenomena leading to cell survival values which deviate from linear-quadratic models. Mutat Res 568: 33–39. [DOI] [PubMed] [Google Scholar]
- Cheng KC, Huang HC, Chen JH, Hsu JW, Cheng HC, Ou CH, Yang WB, Chen ST, Wong CH, Juan HF. 2007. Ganoderma lucidum polysaccharides in human monocytic leukemia cells: from gene expression to network construction. BMC Genomics 8: 411–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrales RM, Galarreta D, Herreras J, Calonge M, Chaves F. 2011. Antioxidant enzyme mRNA expression in conjunctival epithelium of healthy human subjects. Can J Ophthalmol 46: 35–39. [DOI] [PubMed] [Google Scholar]
- Criswell T, Beman M, Araki S, Leskov K, Cataldo E, Mayo LD, Boothman DA. 2005. Delayed activation of insulin-like growth factor-1 receptor/Src/MAPK/Egr-1 signaling regulates clusterin expression, a pro-survival factor. J Biol Chem. 280:14212–14221. [DOI] [PubMed] [Google Scholar]
- Dauer LT, Brooks AL, Hoel DG, Morgan WF, Stram D, Tran P. 2010. Review and evaluation of updated research on the health effects associated with low-dose ionising radiation. Radiat Prot Dosimetry 140: 103–136. [DOI] [PubMed] [Google Scholar]
- Day TK, Zeng G, Hooker AM, Bhat M, Scott BR, Turner DR, Sykes PJ. 2006. Extremely low priming doses of X radiation induce an adaptive response for chromosomal inversions in pKZ1 mouse prostate. Radiat Res 166: 757–766. [DOI] [PubMed] [Google Scholar]
- Ding K-K, Shang Z-F, Hao C, Xu Q-Z, Shen J-J, Yang CJ, Xie YH, Qiao C, Wang Y, Xu LL, Zhou PK. 2009. Induced expression of the IER5 gene by gamma-ray irradiation and its involvement in cell cycle checkpoint control and survival. Radiat Environ Biophys 48: 205–213. [DOI] [PubMed] [Google Scholar]
- Ding L-H, Shingyoji M, Chen F, Hwang J-J, Burma S, Lee C, Cheng JF, Chen DJ. 2005. Gene expression profiles of normal human fibroblasts after exposure to ionizing radiation: a comparative study of low and high doses. Radiat Res 164: 17–26. [DOI] [PubMed] [Google Scholar]
- Etemadmoghadam D, George J, Cowin PA, Cullinane C, Kansara M. Australian Ovarian Cancer Study Group Gorringe KL, Smyth GK, Bowtell DD. 2010. Amplicon-dependent CCNE1 expression is critical for clonogenic survival after cisplatin treatment and is correlated with 20q11 gain in ovarian cancer. PLoS One 5: e15498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinendegen LE, Neumann RD. 2005. Physics must join with biology in better assessing risk from low-dose irradiation. Radiat Prot Dosimetry 117: 346–356. [DOI] [PubMed] [Google Scholar]
- Franco N, Lamartine J, Frouin V, Le Minter P, Petat C, Leplat JJ, Libert F, Gidrol X, Martin MT. 2005. Low-dose exposure to gamma rays induces specific gene regulations in normal human keratinocytes. Radiat Res 163: 623–635. [DOI] [PubMed] [Google Scholar]
- Gaube F, Wolfl S, Pusch L, Kroll TC, Hamburger M. 2007. Gene expression profiling reveals effects of Cimicifuga racemosa (L.) NUTT.(black cohosh) on the estrogen receptor positive human breast cancer cell line MCF-7. BMC Pharmacol 7: 11–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S, Krishna M. 2012. Role of Rad52 in fractionated irradiation induced signaling in A549 lung adenocarcinoma cells. Mutat Res 729: 61–72. [DOI] [PubMed] [Google Scholar]
- Gong B, Almasan A. 2000. Apo2 ligand/TNF-related apoptosis-inducing ligand and death receptor 5 mediate the apoptotic signaling induced by ionizing radiation in leukemic cells. Cancer Res 60: 5754–5760. [PubMed] [Google Scholar]
- Grudzenski S, Raths A, Conrad S, Rübe CE, Löbrich M. 2010. Inducible response required for repair of low-dose radiation damage in human fibroblasts. Proc Natl Acad Sci 107: 14205–14210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinloth AN, Shackelford RE, Innes CL, Bennett L, Li L, Amin RP, Sieber SO, Flores KG, Bushel PR, Paules RS. 2003. ATM-dependent and -independent gene expression changes in response to oxidative stress, gamma irradiation, and UV irradiation. Radiat Res 160: 273–290. [DOI] [PubMed] [Google Scholar]
- Horne MC, Goolsby GL, Donaldson KL, Tran D, Neubauer M, Wahl AF. 1996. Cyclin G1 and cyclin G2 comprise a new family of cyclins with contrasting tissue-specific and cell cycle-regulated expression. J Biol Chem 271: 6050–6061. [DOI] [PubMed] [Google Scholar]
- Jain AK, Allton K, Iacovino M, Mahen E, Milczarek RJ, Zwaka TP, Kyba M, Barton MC. 2012. p53 regulates cell cycle and microRNAs to promote differentiation of human embryonic stem cells. PLoS Biol 10: e1001268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerman IA, Bernard R, Bunney WE, Jones EG, Schatzberg AF, Myers RM, Barchas JD, Akil H, Watson SJ, Thompson RC. 2012. Evidence for transcriptional factor dysregulation in the dorsal raphe nucleus of patients with major depressive disorder. Front Neurosci 6: 135–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Oh J, Choi JY, Jang JY, Kang MW, Lee CE. 2008. Identification of human thioredoxin as a novel IFN-gamma-induced factor: mechanism of induction and its role in cytokine production. BMC Immunol 9: 64–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kis E, Szatmari T, Keszei M, Farkas R, Esik O, Lumniczky K, Falus A, Sáfrány G. 2006. Microarray analysis of radiation response genes in primary human fibroblasts. Int J Radiat Oncol Biol Phys 66: 1506–1514. [DOI] [PubMed] [Google Scholar]
- Kojima S, Matsuki O, Nomura T, Kubodera A, Honda Y, Honda S, Tanooka H, Wakasugi H, Yamaoka K. 1998. Induction of mRNAs for glutathione synthesis-related proteins in mouse liver by low doses of gamma-rays. Biochim Biophys Acta 1381: 312–318. [DOI] [PubMed] [Google Scholar]
- Lew DJ, Dulić V, Reed SI. 1991. Isolation of three novel human cyclins by rescue of G1 cyclin (Cln) function in yeast. Cell 66: 1197–1206. [DOI] [PubMed] [Google Scholar]
- Li H, Wan A, Xu G, Ye D. 2013. Small changes huge impact: the role of thioredoxin 1 in the regulation of apoptosis by S-nitrosylation. Acta Biochim Biophys Sin (Shanghai) 45: 153–161. [DOI] [PubMed] [Google Scholar]
- Marples B, Collis SJ. 2008. Low-dose hyper-radiosensitivity: past, present, and future. Int J Radiat Oncol Biol Phys 70: 1310–1318. [DOI] [PubMed] [Google Scholar]
- Marples B, Joiner MC. 1993. The response of Chinese hamster V79 cells to low radiation doses: evidence of enhanced sensitivity of the whole cell population. Radiat Res 133: 41–51. [PubMed] [Google Scholar]
- Marples B, Wouters BG, Joiner MC. 2003. An association between the radiation-induced arrest of G2-phase cells and low-dose hyper-radiosensitivity: a plausible underlying mechanism? Radiat Res 160: 38–45. [DOI] [PubMed] [Google Scholar]
- Matsumoto H, Hamada N, Takahashi A, Kobayashi Y, Ohnishi T. 2007. Vanguards of paradigm shift in radiation biology: radiation-induced adaptive and bystander responses. J Radiat Res 48: 9097–9106. [DOI] [PubMed] [Google Scholar]
- Mezentsev A, Amundson SA. 2011. Global gene expression responses to low- or high-dose radiation in a human three-dimensional tissue model. Radiat Res 175: 677–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzayans R, Andrais B, Scott A, Wang YW, Murray D. 2013. Ionizing Radiation-Induced Responses in Human Cells with Differing TP53 Status. Int J Mol Sci 14: 22409–22435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchel RE, Burchart P, Wyatt H. 2008. A lower dose threshold for the in vivo protective adaptive response to radiation.Tumorigenesis in chronically exposed normal and Trp53 heterozygous C57BL/6 mice. Radiat res 170: 765–775. [DOI] [PubMed] [Google Scholar]
- Morgan WF, Bair WJ. 2013. Issues in low dose radiation biology: the controversy continues.A perspective. Radiat Res 179: 501–510. [DOI] [PubMed] [Google Scholar]
- Moskalev AA, Plyusnina EN, Shaposhnikov MV. 2011. Radiation hormesis and radioadaptive response in Drosophila melanogaster flies with different genetic backgrounds: the role of cellular stress-resistance mechanisms. Biogerontology. 12: 253–263. [DOI] [PubMed] [Google Scholar]
- Nasonova EA, Shmakova NL, Komova OV, Mel’nikova LA, Fadeeva TA, Krasavin EA, Ritter S. 2006. Cytogenetic effects of low-dose radiation with different LET in human peripheral blood lymphocytes. Radiat Environ Biophys 45: 307–312. [DOI] [PubMed] [Google Scholar]
- Neumaier T, Swenson J, Pham C, Polyzos A, Lo AT, Yang P, Dyball J, Asaithamby A, Chen DJ, Bissell MJ, Thalhammer S, Costes SV. 2012. Evidence for formation of DNA repair centers and dose-response nonlinearity in human cells. Proc Natl Acad Sci 109: 443–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosel I, Vaurijoux A, Barquinero JF, Gruel G. 2013. Characterization of gene expression profiles at low and very low doses of ionizing radiation. DNA Repair (Amst) 12: 508–517. [DOI] [PubMed] [Google Scholar]
- Obtulowicz T, Swoboda M, Speina E, Gackowski D, Rozalski R, Siomek A, Janik J, Janowska B, Ciesla JM, Jawien A, Banaszkiewicz Z, Guz J, Dziaman T, Szpila A, Olinski R, Tudek B. 2010. Oxidative stress and 8-oxoguanine repair are enhanced in colon adenoma and carcinoma patients. Mutagenesis 25: 463–471. [DOI] [PubMed] [Google Scholar]
- Ohtsuka T, Jensen MR, Kim HG, Kim K-T, Lee SW. 2004. The negative role of cyclin G in ATM-dependent p53 activation. Oncogene 23: 5405–5408. [DOI] [PubMed] [Google Scholar]
- Osipov AN, Buleeva G, Arkhangelskaya E, Klokov D. 2013. In vivo gamma-irradiation low dose threshold for suppression of DNA double strand breaks below the spontaneous level in mouse blood and spleen cells. Mutat Res 756: 141–145. [DOI] [PubMed] [Google Scholar]
- Osipov AN, Klokov DY, Elakov AL, Rozanova OM, Zaichkina SI, Aptikaeva GF, Akhmadieva AKh. 2004. Comparison in vivo study of genotoxic action of high- versus very low dose-rate gamma-irradiation. Nonlinearity Biol Toxicol Med 2: 223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plyusnina EN, Shaposhnikov MV, Moskalev AA. 2011. Increase of Drosophila melanogaster lifespan due to D-GADD45 overexpression in the nervous system. Biogerontology 12: 211–226. [DOI] [PubMed] [Google Scholar]
- Pollycove M, Feinendegen LE. 2003. Radiation-induced versus endogenous DNA damage: possible effect of inducible protective responses in mitigating endogenous damage. Hum Exp Toxicol 22: 290–306; discussion 307, 315–297, 319–223. [DOI] [PubMed] [Google Scholar]
- Prasad AV, Mohan N, Chandrasekar B, Meltz ML. 1995. Induction of transcription of “immediate early genes” by low-dose ionizing radiation. Radiat Res 143: 263–272. [PubMed] [Google Scholar]
- Ramana CV, Boldogh I, Izumi T, Mitra S. 1998. Activation of apurinic/apyrimidinic endonuclease in human cells by reactive oxygen species and its correlation with their adaptive response to genotoxicity of free radicals. Proc Natl Acad Sci 95: 5061–5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaud E, Miccoli L, Zacal N, Biard DS, Craescu CT, Rainbow AJ, Angulo JF. 2011. Differential contribution of XPC, RAD23A, RAD23B and CENTRIN 2 to the UV-response in human cells. DNA Repair (Amst) 10: 835–847. [DOI] [PubMed] [Google Scholar]
- Riches LC, Lynch AM, Gooderham NJ. 2010. A molecular beacon approach to detecting RAD52 expression in response to DNA damage in human cells. Toxicol In Vitro 24: 652–660. [DOI] [PubMed] [Google Scholar]
- Rothkamm K, Löbrich M. 2003. Evidence for a lack of DNA double-strand break repair in human cells exposed to very low x-ray doses. Proc Natl Acad Sci 100: 5057–5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy N, Stoyanova T, Dominguez-Brauer C, Park HJ, Bagchi S, Raychaudhuri P. 2010. DDB2, an essential mediator of premature senescence. Mol Cell Biol 30: 2681–2692. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sadekova S, Lehnert S, Chow TY. 1997. Induction of PBP74/mortalin/Grp75, a member of the hsp70 family, by low doses of ionizing radiation: a possible role in induced radioresistance. Int J Radiat Biol 72: 653–660. [DOI] [PubMed] [Google Scholar]
- Saha MN, Jiang H, Yang Y, Zhu X, Wang X, Schimmer AD, Qiu L, Chang H. 2012. Targeting p53 via JNK pathway: a novel role of RITA for apoptotic signaling in multiple myeloma. PLoS One 7: e30215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaposhnikov MV, Moskalev AA, Plyusnina EN. 2011. Effect of PARP-1 overexpression and pharmacological inhibition of NF-kB on the lifespan of Drosophila melanogaster. Adv Gerontol 24: 405–419. [PubMed] [Google Scholar]
- Sharma AK, Ali A, Gogna R, Singh AK, Pati U. 2009. p53 Amino-terminus region (1–125) stabilizes and restores heat denatured p53 wild phenotype. PLoS One 4: e7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Słonina D, Biesaga B, Urbanski K, Kojs Z, Waligórski M. 2006. Evidence of low-dose hyper-radiosensitivity in normal cells of cervix cancer patients? Radiat Prot Dosimetry 122: 282–284. [DOI] [PubMed] [Google Scholar]
- Tice RR, Agurell E, Anderson D, Burlinson B, Hartmann A, Kobayashi H, Miyamae Y, Rojas E, Ryu JC, Sasaki YF. 2000. Single cell gel/comet assay: guidelines for in vitro and in vivo genetic toxicology testing. Environ Mol Mutagen 35: 206–221. [DOI] [PubMed] [Google Scholar]
- UNSCEAR (United Nations Scientific Committee on the Effects of Atomic Radiation). 2012. Biological mechanisms of radiation action at low doses.V.12-57831.United Nations, New York, USA. [Google Scholar]
- Xu B, Kim S-T, Lim D-S, Kastan MB. 2002. Two molecularly distinct G(2)/M checkpoints are induced by ionizing irradiation. Mol Cell Biol 22: 1049–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X-X, Liu C-B, Chen J-Y, Tao B-H, Zhi-Yi C. 2012. The expression of cyclin G in nasopharyngeal carcinoma and its significance. Clin Exp Med 12: 21–24. [DOI] [PubMed] [Google Scholar]
- Zhao L, Samuels T, Winckler S, Korgaonkar C, Tompkins V, Horne MC, Quelle DE. 2003. Cyclin G1 has growth inhibitory activity linked to the ARF-Mdm2-p53 and pRb tumor suppressor pathways. Mol Cancer Res 1: 195–206. [PubMed] [Google Scholar]
- Zimmermann M, Arachchige-Don AS, Donaldson MS, Dallapiazza RF, Cowan CE, Horne MC. 2012. Elevated cyclin G2 expression intersects with DNA damage checkpoint signaling and is required for a potent G2/M checkpoint arrest response to doxorubicin. J Biol Chem 287: 22838–22853. [DOI] [PMC free article] [PubMed] [Google Scholar]