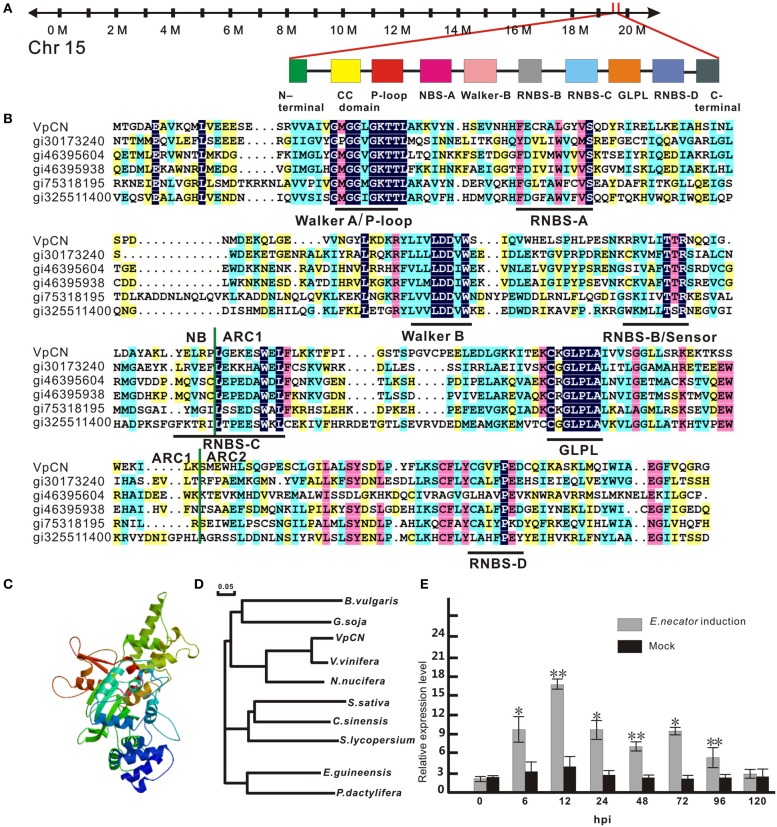
Figure 1.
Sequence analysis of VpCN and transcript level detection. (A) Schematic map of VpCN location and major motifs. (B) Multiple sequence alignment of the NB, ARC1 and ARC2 subdomains of NB-ARC in VpCN with closely related proteins. Domain borders are indicate as vertical green lines. Motifs are labeled by horizontal dark lines below the aligned sequences. gi30173240 (Bent et al., 1994), gi46395604 (Bevan et al., 1998), gi46395938 (Theologis et al., 2000), gi75318159 (Ori et al., 1997), gi325511400 (Theologis et al., 2000) (C) Structural model of the NB-ARC domain of VpCN. (D) Phylogenetic tree of VpCN and related proteins from other plant species. The tree was generated using the ClustalW function in the MegAlign program: Vitis vinifera (GenBank accession no. XP010661747), Nelumbo nucifera (GenBank accession no. XP0102588251), Glycine soja (GenBank accession no. KHN19144), Elaeis guineensis (GenBank accession no. XP010913221), Solanum lycopersicum (GenBank accession no. XP010319316), Beta vulgaris subsp. Vulgaris (GenBank accession no. XP010669409), Phoenix dactylifera (GenBank accession no. XP008791188), Camelina sativa (GenBank accession no. XP010426119), Citrus sinensis (GenBank accession no. XP006470644). The scale bar represents 0.05 substitutions per site. (D) Structure model of NB-ARC in VpCN. (E) Analysis of VpCN expression in response to E. necator inoculation. The third to fifth fully expanded young grapevine leaves beneath the apex were selected for samples. The experiment encompass three independent biological replicates, for each biological replicate three leaves haversted from three plant and three technical replicates were performed. Data represent means of three biological replicates ±SE, asterisksin indicate statistical significance in comparison with control (Student'st-test, significance levels of *P < 0.05, **P < 0.01 are indicated).