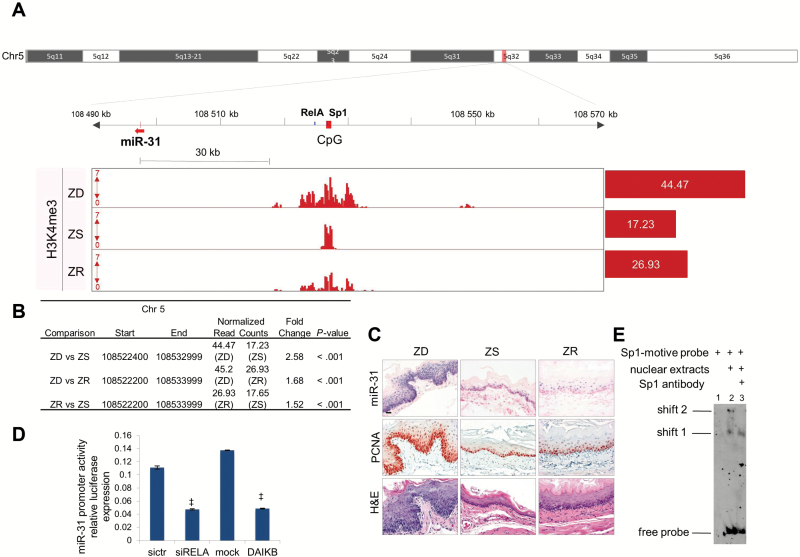
Figure 7.
ChIP-seq identification of the promoter region of miR-31 and TFBs. A) Integrative Genomics Viewer view shows the putative miR-31 promoter region in Zn-deficient (ZD) rat esophagus: miR-31 (red arrow), CpG island (red box), and TFBs NF-κΒ/Sp1 (blue box). The values on the y-axis for ChIP-seq data are input-normalized intensities; the values on red barplots are normalized read counts/library size x 1 000 000. B) Table shows differentially enriched regions (H3K4me3) among Zn-modulated groups. C) Hematoxylin and eosin stain analysis of histology, in situ hybridization analysis of miR-31 expression, and immunohistochemical analysis of the proliferation marker PCNA of Zn-modulated esophagus (scale bars = 25 μm). D) NF-κΒ transcriptionally activates miR-31. Luciferase experiment shows downregulation of miR-31 promoter activity after NF-κΒ silencing or the overexpression of a NF-κΒ–dominant active inhibitor (experiments were performed three times, ‡P < .001, error bars = ± standard deviation). All statistical tests were two-sided. E) DNA mobility shift assay shows the binding of Sp1 to Sp1-probe motive. Lane 1: labeled Sp1-motive probe. Lane 2: two complexes at different molecular weights (shift 1, shift 2) are formed by the addition of nuclear extracts from HELA cells to Sp1-motive probe. Lane 3: anti-Sp1 antibody disrupts the higher molecular weight complex (shift 2). DAIKB = NF-κΒ–dominant active inhibitor; siRELA = NF-κΒ silencing; ZD = Zn-deficient; ZR = Zn-replenished; ZS = Zn-sufficient.