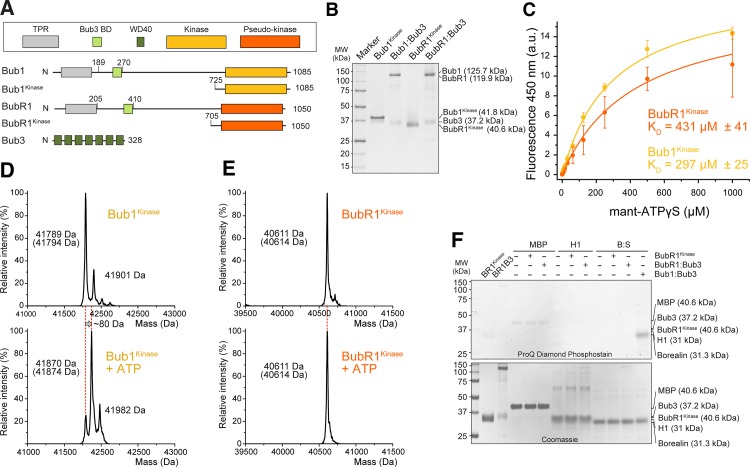
Fig 1. Reconstitution of Bub1 kinase and BubR1 pseudo-kinase.
(A) Bub1 and BubR1 share a similar domain organization. Schematic description of the domains and constructs used in this manuscript: TPR–tetratricopeptide repeat, BD–binding domain, WD40 –an approximately 40-residue sequence repeat often terminating with a tryptophan (W) and aspartate (D) dipeptide. (B) Purified Bub1kinase, Bub1:Bub3, BubR1kinase, and BubR1:Bub3 were separated by SDS-PAGE. Their respective expected molecular mass is indicated. (C) Both Bub1kinase and BubR1kinase bind to mant-ATP. The change in fluorescence emission at 450 nm is plotted as a function of total mant-ATP concentration. The data were fitted with a one site binding equation using Origin 9.0, with R2 = 0.99 for both curves. Error bars represent SD of a mean of at least 2 independent experiments. a.u.–arbitrary units. (D-E) ESI-MS spectra of purified Bub1kinase (D) or BubR1kinase (E) before and after incubation with ATP for auto-phosphorylation. Theoretical calculated masses are given in brackets under the measured masses. (F) BubR1 is not an active kinase. Maltose binding protein (MBP), H1, and the Borealin:Survivin complex (Bor:Sur) were incubated with 50 nM BubR1 constructs and ATP and analyzed on SDS PAGE visualizing phosphates using the Pro-Q® Diamond Phosphoprotein Gel Stain. 10 nM Bub1:Bub3 was used as a positive control. BR1kinase–BubR1kinase, BR1B3 –BubR1:Bub3.