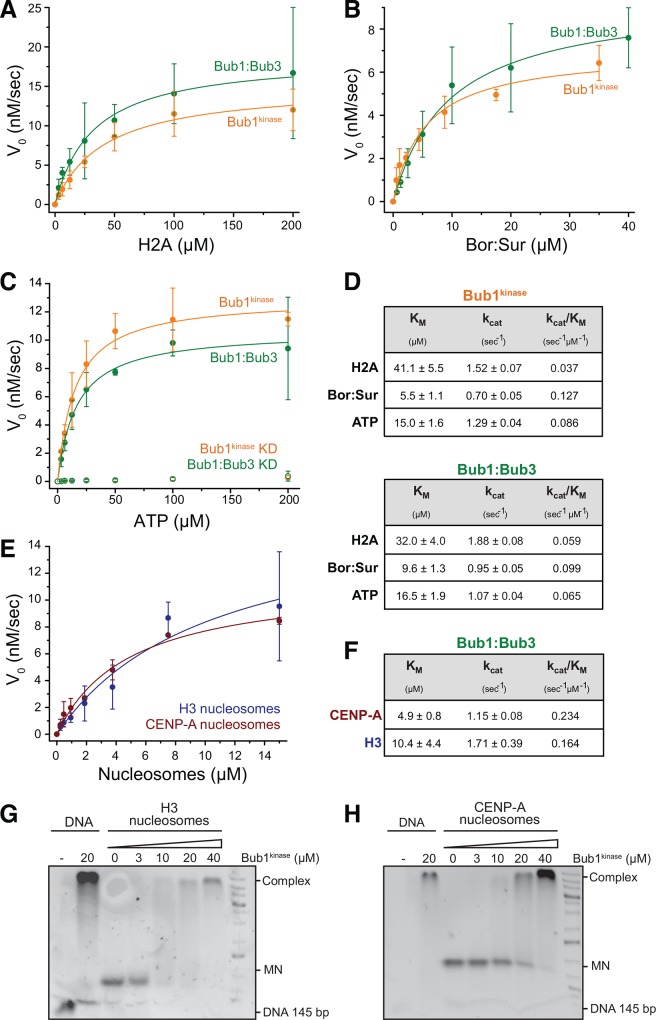
Fig 2. Kinetic characterization of Bub1 complexes.
(A-C) Bub1kinase and Bub1:Bub3 complex exhibit similar catalytic activity toward H2A (A) and Bor:Sur substrates (B) and hydrolyze ATP at similar rates (C). The mutation D917N abrogates ATPase activity (C). The kinase activity was determined using the ADP-GloTM Kinase Assay and is plotted as a function of substrate concentration to allow fitting according to the Michaelis-Menten equation with R2 = 0.99 (Bub1kinase on Bor:Sur R2 = 0.97). KD–kinase dead. Error bars represent SD of a mean of at least 2 independent experiments. (D) Kinetic parameters of the Michaelis-Menten fits as determined in (A-C). (E) H2A contained in H3- or CENP-A nucleosomes is efficiently phosphorylated by Bub1kinase and Bub1:Bub3. (F) The kinase activity, plotted as a function of substrate concentration, allows fitting with the Michaelis-Menten equation with R2 = 0.95 (H3) and 0.99 (CENP-A). Error bars represent SD of a mean of at least two independent experiments. (G-H) EMSA assays showing DNA and nucleosome binding of Bub1kinase and H3- or CENP-A nucleosomes. MN–mononucleosomes.