Abstract
The use of microorganisms as cell factories frequently requires extensive molecular manipulation. Therefore, the identification of genomic neutral sites for the stable integration of ectopic DNA is required to ensure a successful outcome. Here we describe the genome mapping and validation of five neutral sites in the chromosome of Synechocystis sp. PCC 6803, foreseeing the use of this cyanobacterium as a photoautotrophic chassis. To evaluate the neutrality of these loci, insertion/deletion mutants were produced, and to assess their functionality, a synthetic green fluorescent reporter module was introduced. The constructed integrative vectors include a BioBrick-compatible multiple cloning site insulated by transcription terminators, constituting robust cloning interfaces for synthetic biology approaches. Moreover, Synechocystis mutants (chassis) ready to receive purpose-built synthetic modules/circuits are also available. This work presents a systematic approach to map and validate chromosomal neutral sites in cyanobacteria, and that can be extended to other organisms.
Keywords: neutral sites, photoautotrophic chassis, Synechocystis, synthetic biology
1. Introduction
Microorganisms have been widely used in biotechnological applications to produce added-value products.1 The Gram-negative bacterium Escherichia coli is the most commonly used, but others such as the Gram-positive bacterium Bacillus subtilis and the eukaryotic yeast Saccharomyces cerevisiae have also been successfully exploited.2,3 Heterotrophic organisms require supplementation of the growth medium with external carbon sources increasing production costs; therefore, autotrophic organisms emerge as a valid alternative.4 In this context, cyanobacteria are promising ‘low-cost’ cell factories since they can use CO2 as carbon source, water as reducing power, light as energy source and some strains are even able to fix atmospheric N2 (nitrogen source). These organisms are found in almost any ecological niche on Earth,5 which reflects their high degree of metabolic plasticity. Among cyanobacteria, the unicellular non-N2-fixing Synechocystis sp. PCC 6803 (henceforth referred to as Synechocystis) is the most studied strain, and the large amount of genomic, proteomic and physiological information generated over the years allowed the construction of genome-scale models, enabling the prediction of the system behavior.6 Moreover, it was the first photosynthetic organism to have its genome sequenced,7 comprising a 3.6 Mb chromosome and 7 plasmids (http://genome.microbedb.jp/cyanobase/Synechocystis), it is naturally transformable and a number of molecular tools are available for its genetic manipulation.4,8,9 Altogether, these characteristics allow for the exploitation of Synechocystis as a photoautotrophic biotechnological platform. In contrast to E. coli, for which several replicative plasmids from different compatibility groups are available, these tools are limited for Synechocystis.8–10 Instead, integrative vectors are usually used to implement new functionalities, integrating ectopic DNA into the genome through homologous recombination.11 To date, a few integration sites in Synechocystis' genome have been used,11–15 although most of them have not been extensively characterized as neutral sites—loci that can be disrupted without affecting cellular viability or cause any distinguishable phenotype.16 Therefore, a comprehensive and systematic approach to identify and characterize genomic neutral sites would greatly improve the ability to introduce new functionalities into Synechocystis.
In this work, a set of neutral sites in the chromosome of Synechocystis has been identified and characterized. It presents a systematic approach to map and validate neutral sites in the genome of an organism, foreseeing its use as a photoautotrophic chassis for industrial applications. Moreover, the new integrative vectors have been designed to be compatible with the BioBrick™ RFC[10] standard (http://www.biobricks.org/), which will make easier to use Synechocystis in synthetic biology approaches. The availability of several integration loci, spread throughout the cyanobacterium chromosome, will allow the implementation of complex synthetic circuits into this platform. Furthermore, the strategy reported here can be easily extended to other organisms/chassis.
2. Materials and methods
2.1. Organisms and maintenance culture conditions
Wild type and mutants of the cyanobacterium Synechocystis sp. PCC 6803 substrain Kazusa17,18 were maintained in BG11 medium19 at 25°C, under a 16 h light/8 h dark regimen. Light intensity was 20 µmol photons m−2 s−1 in all experiments. For solid medium, BG11 was supplemented with 1.5% (wt/vol) noble agar (Difco), 0.3% (wt/vol) sodium thiosulfate and 10 mM TES-KOH buffer, pH 8.2. For the selection and maintenance of mutants, BG11 medium was supplemented with kanamycin (Km, 10–600 µg ml−1). All the characterization experiments were performed in the absence of selective pressure. For cloning purposes, E. coli strain DH5α (Stratagene) was used. Escherichia coli cells were grown at 37°C on LB medium,20 supplemented with ampicillin (100 µg ml−1) or kanamycin (50 µg ml−1).
2.2. Bioinformatics
The list of the predicted/annotated ORFs of Synechocystis was retrieved from CyanoBase (Distribution of Sequence and Annotated Data Files, ftp://ftp.kazusa.or.jp/pub/CyanoBase/Synechocystis, accessed: 08 May 2009). From the 3,264 ORFs listed, only those putatively encoding unknown or hypothetical proteins were selected. From these, the final selection was based on the following criteria: (i) length of the putatively encoded proteins ≤301 amino acids, (ii) encoded proteins with no transmembrane domains predicted by the TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/), (iii) no interaction with other proteins as assessed by the two-hybrid system (CyanoBase data) and (iv) no relevant similarities found at the protein sequence level, when comparing with other sequences using BLASTP (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The genomic context of the ORFs was also taken into consideration, and sites were disregarded when in the vicinity of genes with assigned putative functions (see Fig. 1). To assess transcription of the selected ORFs, primers were designed automatically from the genomic sequence7 using the Beacon Designer 6 software (PREMIER Biosoft International). For other purposes, primers were designed manually and analysed using the Integrated DNA Technologies web resource OligoAnalyzer v3.1 (http://eu.idtdna.com/analyzer/Applications/OligoAnalyzer/).
Figure 1.

Chromosomal location and genomic context of the Synechocystis' putative neutral sites. (A) Green, sites chosen for mutants construction, Red, sites that fulfil the neutral sites initial selection criteria, but that were subsequently disregarded after additional analyses. (B) Genomic context of each putative neutral site (5,000 bp). Blue, ORFs corresponding to the neutral sites, Red, ORFs encoding proteins with assigned functions; tp, putative transposase; ‘, partial ORFs.
2.3. DNA and RNA extraction
Cyanobacterial genomic DNA (gDNA) extraction was carried out according to the procedure described previously.21 For RNA extraction, Synechocystis cells were collected by centrifugation (10 min at 3,850 g) and frozen at −80°C. RNA was extracted using the TRIzol® reagent (Invitrogen) according to the method described previously.22
2.4. RT-PCR for target ORFs transcription analysis
To assess target ORFs transcription, RNA was extracted from Synechocystis cells cultivated to an OD730 of 0.8–0.9, under continuous light at 30°C. Further transcriptional studies were performed with RNA extracted from samples collected in three different growth phases (OD730 of ∼0.4, 2 and 9, see Fig. 2), from three independent cultures grown under the same conditions. RT-PCRs were carried out as described elsewhere.23 Briefly, cDNA was synthesized in duplicate from 1 µg of total RNA using the iScript™ Select cDNA Synthesis Kit (Bio-Rad) and the random primers supplied, following the manufacturer's instructions. The two RT reactions from each sample were pooled together and each PCR reaction contained 1.5 µl of cDNA, 0.1 µM of each primer (Supplementary Table S1), 200 µM dNTPs mix, 1× PCR reaction buffer and 0.025 U of illustra™ Taq DNA polymerase (GE Healthcare). The PCR profile was as follows: 2 min at 94°C followed by 40 cycles of 30 s at 94°C, 30 s at 50°C and 30 s at 72°C, concluding with a 7-min extension step at 72°C. RNA contamination by gDNA prior to cDNA synthesis was assessed by PCR using primers specific for the Synechocystis 16S rDNA, and the RT reaction efficiency was assessed using the same primers pair. All the RT-PCRs included a negative control (omission of template) and a positive control (Synechocystis gDNA).
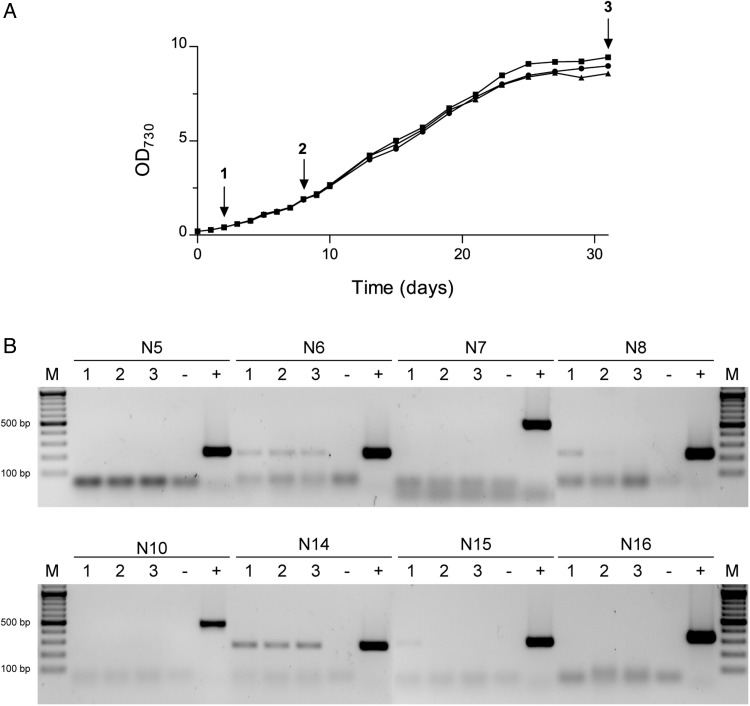
Figure 2.
Synechocystis growth curves, sample collection and transcription analysis of eight selected loci (N5, N6, N7, N8, N10, N14, N15 and N16). (A) Growth curves of three independent cultures of Synechocystis with sample collection points for RNA extraction indicated by arrows (1—OD730 ≈ 0.4; 2—OD730 ≈ 1.9; 3—OD730 ≈ 9.0). (B) RT-PCR transcription analysis using total RNA extracted from cells collected at the time points indicated in A. These results are representative of three biological replicates and technical duplicates. −, negative control (absence of template); +, positive control (genomic DNA); and M, GeneRuler DNA ladder (Thermo Fisher Scientific Inc.).
2.5. Homologous regions amplification and fusion by PCR
PCRs to amplify the regions flanking the target loci (see below) contained 7 ng of gDNA, 0.2 µM of each primer, 250 µM dNTPs mix, 1× Pfu reaction buffer with MgSO4 and 0.025 U of Pfu DNA polymerase (Thermo Fisher Scientific Inc.). The PCR profile was as follows: 1 min at 94°C followed by 35 cycles of 30 s at 94°C, 45 s at 48°C and 90 s at 72°C, concluding with a 7-min extension step at 72°C. Agarose gel electrophoresis was performed by standard protocols using 1× TAE buffer;20 DNA fragments were purified using the GFX™ PCR DNA and Gel Band Purification Kit (GE Healthcare), according to the manufacturer's instructions. ‘Overlap-PCRs’ (see below) contained 80 ng of each purified DNA fragment, 0.125 µM of each primer, 250 µM dNTPs mix, 1× PCR reaction buffer and 0.025 U of illustra™ Taq DNA polymerase (GE Healthcare). The PCR profile was: 5 min at 95°C followed by 10 cycles of 30 s at 95°C, 45 s at 48°C and 90 s at 72°C, and 30 cycles of 30 s at 95°C, 45 s at 55°C and 90 s at 72°C, concluding with a 7-min extension step at 72°C. The oligonucleotide primers used to amplify the homologous regions are listed in Supplementary Table S2.
2.6. Plasmid construction
The plasmids used to produce the Synechocystis mutants were constructed following a rational design (Supplementary Fig. S1). For this purpose, the chromosomal regions up and downstream of the target loci (Supplementary Table S3) were amplified by PCR using the Pfu polymerase and primers containing tags with restriction sites for cloning purposes (Nn.5O/Nn.5I and Nn.3O/Nn.3I, Supplementary Table S2). The corresponding up and downstream fragments of each locus were fused by ‘overlap-PCR’, using the Taq polymerase (see above). The resulting products were cloned into the pGEM®-T easy vector (Promega), according to the manufacturer's instructions, originating the pSNn* plasmids series (n for ‘neutral site number’). The pGEM®-T easy vector restriction sites incompatible with the BioBrick RFC[10] standard were eliminated by digesting the vector with XhoI and SalI and recircularizing the vector (originating the pSNn** series). The BioBrick RFC[10] standard incompatible restriction sites within the amplified chromosomal regions of pSN5** and pSN10** were eliminated using the QuikChange® Multi Site-Directed Mutagenesis Kit (Stratagene), according to the manufacturer's instructions, using the primers N5_SDM and N10_SDM, respectively (Supplementary Table S2), originating the pSN5*** and the pSN10*** plasmids. A BioBrick-compatible cloning interface was synthesized (Epoch Life Science Inc.), containing the MunI, XbaI, NotI, SpeI and PstI restriction sites flanked by two double transcription terminators (based on the registry parts BBa_B1006, BBa_B0062, BBa_B0053 and BBa_B0020). This interface was excised from the delivery vector (pBSK-FP300) using XmaI and AgeI and subsequently cloned into the XmaI site of the pSNn** or pSNn*** plasmids, originating the pSNn plasmid series (Supplementary Fig. S1). Afterwards, two selection cassettes, one containing the nptII gene (encoding a neomycin phosphotransferase that confers resistance to neomycin and kanamycin) and the sacB gene (encoding a levansucrase that confers sucrose sensitivity), and another containing only the nptII gene, were amplified by PCR from the plasmid pK18mobsacB,24 using the primer pairs Km.KmScFwd/KmScRev and Km.KmScFwd/KmRev, respectively (Supplementary Table S2). These selection cassettes were cloned into the pGEM-T easy vector, excised with XmaI and cloned into the XmaI site of the pSNn plasmid series. The final plasmid series containing only the Km-resistance cassette was named pSNnK, and the plasmid series containing the cassette with the two selective markers was named pSNnKS. Additionally, based on previous works,25,26 the minimal promoter sequence required for Synechocystis psbA2 basal transcription (−38 to +14 bp (base-pairs); +1 corresponding to the transcription start site) was synthesized (PpsbA2*), meeting the BioBrick RFC[10] standard specifications (DNA 2.0 Inc.). This promoter region was cloned into the BioBrick vector pSB1A2 containing the part BBa_E0240 (composed by the RBS part BBa_B0032, the GFP-coding gene part BBa_E0040 and the double transcription terminator part BBa_B0015), according to the BioBrick standard cloning procedure. The BioBrick composite part BBa_E0240 (promoterless GFP generator) and the part with the synthetic constitutive promoter PpsbA2* were digested with XbaI and SpeI and cloned into the XbaI site of both pSNn and pSNnK series, originating the pSNnK.gfp and pSNnK.Cgfp plasmid series (C for ‘constitutive promoter’). These plasmids were used to assess proper cloning interface transcriptional insulation (promoterless gfp) and functionality of the neutral sites (gfp under the control of the constitutive promoter). The vectors were named as follows: P for ‘plasmid’, S for ‘Synechocystis’, N for ‘neutral site’, n for the ‘number of the neutral site’ (5, 8, 10, 15 or 16), K for ‘kanamycin resistance cassette’, KS for the ‘double selection cassette (KmR/SucS)’, C for ‘constitutive PpsbA2* promoter’ and gfp for the ‘GFP generator BioBrick part BBa_E0240’. Plasmids DNA was isolated from overnight grown E. coli cultures, using the GenElute™ Plasmid Miniprep Kit (Sigma-Aldrich), according to the manufacturer's instructions. All the BioBrick parts can be found at the Registry of Standard Biological Parts website (http://parts.igem.org), and the maps and sequences (FASTA format) of the final plasmids are available as Supplementary Figs S2–S26.
2.7. Generation of Synechocystis mutants
Synechocystis was transformed based on the procedure described previously.11 Briefly, Synechocystis was grown in standard conditions to an OD730 ≈ 0.5. Cells were harvested by centrifugation (10 min at 3,850 g) and resuspended in BG11 to a final OD730 ≈ 2.5. The purified plasmids were incubated for 5 h with 500 µl of those cells (final plasmid DNA concentration of 20 µg ml−1), in light and at 25°C. Cells were then spread onto Immobilon™-NC membranes (0.45 µm pore size, 82 mm, Millipore) resting on solid BG11 plates, grown in standard growth conditions and transferred to selective plates after 24 h (Km, 10 µg ml−1). Transformants were observed after 1–2 weeks. For complete segregation, Km-resistant colonies were grown at increasing Km concentrations (25 and 50 µg ml−1) and finally transferred into liquid medium. The mutant full segregation was confirmed by PCR, using primers within and external to each site (Nn.FI/Nn.RI or Nn.FO/KmRev, respectively; Supplementary Table S2), and by Southern blot with probes labeled with digoxigenin using the DIG DNA labeling kit (Roche Molecular Biochemicals) and following the manufacturer's instructions. The probes were obtained by labeling the PCR-amplified 3′ homologous regions for mutants in sites N5, N8, N10 and N16, and the 5′ homologous region for mutants in site N15 (Supplementary Table S2). gDNA digestions were performed with MunI, with the exception of N8 mutants for which the gDNA was digested with SpeI. When necessary antibiotic concentration was increased up to 600 µg ml−1 to fully segregate the mutants.
2.8. Growth experiments
For the growth experiments, 50 ml of BG11 medium was inoculated with Synechocystis wild type or SNnK mutants to OD730 ≈ 0.3. The cultures were grown to OD730 ≈ 1.0, under the experimental light regimen (continuous light or 12 h light/12 h dark cycles) and subsequently diluted to a final OD730 ≈ 0.1 in fresh BG11. Ten millilitres of each diluted culture was transferred to sterile 25-ml borosilicate glass Erlenmeyer flasks, closed with cellulose stopper, and when necessary, glucose was added to a final concentration of 5 mM. The flasks were incubated at constant temperature (30°C) and the indicated light regimens. All experiments were performed in triplicate, and the OD730 of the cultures was recorded daily for 10 consecutive days.
2.9. Statistical analysis of cyanobacteria-specific growth
The quantitative analysis of Synechocystis wild type and neutral sites mutant growth in various conditions was based on the optical density (OD) measurements (see above). For each bin, i, a specific growth parameter (µ), representing the exponent of exponential growth within the given time interval was calculated as:
where ti is time, in units of days running from Day 0 to ti (max) and xi is the corresponding measured OD. As test duration was 10 days in all strains and stationary growth was reached at similar moments, the specific growth rate of all the exponential phase of each replica of each organism was considered to be a reliable parameter to assess growth patterns. ANOVA statistical analysis of the growth data was performed to compare the wild type with each mutant organism, using the Wolfram's Mathematica© software. This ANOVA was three-way where a model with all the effects and interactions up to the third order was tested. Values of up to second-order interactions can be studied in Supplementary Table S4; third-order interactions were in accordance to these results.
2.10. RT-PCR and RT-qPCR for gfp transcription analysis
For total RNA extraction, 400 ml of BG11 medium was inoculated to an OD730 ≈ 0.2 with pre-cultures of Synechocystis wild type and mutants harboring the gfp gene (SNnK.gfp and SNnK.Cgfp mutants' series). Three independent biological replicates of each strain were grown to an OD730 ≈ 1, at 28°C in glass gas washing bottles with aeration, under continuous light. Cells from 100 ml of culture were collected by centrifugation (10 min at 3,850 g); pellets were treated with RNAprotect Bacteria Reagent (Qiagen) and stored at −80°C. Total RNA was extracted as described previously27 and stored at −80°C until further analysis. RNA quality and contamination verifications, cDNA synthesis, RT-PCR and RT-qPCR experiments were performed as described27 using primers listed in Supplementary Table S2. The genes 16S, petB and rnpB were amplified using the primers described elsewhere27 and used as reference genes for data normalization. All RT-qPCR parameters in these experiments were in agreement with the MIQE (minimum information for publication of quantitative real-time PCR experiments) guidelines.28 RT-qPCR data from three biological replicates and three technical replicates were analysed using qbase+ v2.6 (Biogazelle), and the statistical analysis was performed by means of a one-way ANOVA, using GraphPad Prism v6 (GraphPad software Inc.).
2.11. Western blot
For the western blot, 20 ml of the same cultures used for gfp transcription analyses (by RT-PCR and RT-qPCR) was collected by centrifugation (10 min at 3,850 g) and stored at −80°C until further use. Cells were resuspended in lysis buffer (10 mM HEPES pH 8.0, 10 mM EDTA pH 8.0, 0.5% (vol/vol) Triton X-100, 2 mM DTT) supplemented with a cocktail of protease inhibitors (cOmplete Mini, EDTA free, Roche) and disrupted by sonication on ice with a Branson Sonifier 250 using 5 cycles of 15 s (50% dutty cycle, Output 3) intercalated with 1 min off duty. Cell debris was separated by two rounds of 10 min centrifugation at 10,000 g and 4°C. Protein extracts were stored at −20°C until further analysis. Protein quantification was performed using the BCA Protein Assay Kit (Thermo Fisher Scientific Inc.), and 15 µg of each extract was separated by electrophoresis on 12% (wt/vol) SDS-polyacrylamide gels. Protein extracts were visualized with colloidal Coomassie blue staining (Sigma) or transferred to a Hybond™-C extra nitrocellulose membrane (GE Healthcare). After the transfer, membranes were blocked for 3 h with 5% (wt/vol) milk powder in TTBS [Tris-buffered saline (TBS) with 0.05% (vol/vol) Tween 20] and then incubated overnight at 4°C with GFP-specific monoclonal mouse antibody (Roche) in fresh blocking buffer. After washing with TTBS, blots were incubated for 1 h with goat-anti-mouse IgG (Invitrogen) linked to horseradish peroxidase. Membranes were washed with TTBS before immunodetection, which was performed by chemiluminescence using ECL Western Blotting Analysis System detection reagents.
2.12. Confocal microscopy
Ten millilitres of BG11 medium was inoculated with Synechocystis wild type or mutants harbouring the gfp gene (SNnK.gfp and SNnK.Cgfp mutants' series) to OD730 ≈ 0.15. The cultures were grown to OD730 ≈ 1.0, under continuous light at 30°C and subsequently diluted to a final OD730 ≈ 0.1 in fresh BG11. Ten millilitres of each diluted culture was incubated under the same light and temperature conditions as above and grown until OD730 ≈ 1.0. Two hundred microlitres of each culture was collected by centrifugation, resuspended in 20 µl of fresh BG11 and 40 µl of 1% (wt/vol) low-melting point agarose dissolved in BG11 medium was added. Ten microlitres of each mixture was immediately loaded on a glass slide and covered with a coverslip. The GFP emission (collected between 500 and 540 nm) was observed when the cells were exposed to a laser beam at 488 nm, using a Leica TCS SP5 II confocal microscope (Leica Microsystems). Cyanobacterial autofluorescence was collected between 640 and 680 nm after excitation at 633 nm. Images were analysed with the LAS AF Lite v3.3 software (Leica Microsystems). GFP fluorescence measurements were also performed for these samples as described below.
2.13. GFP fluorescence measurements
Fifty millilitres of BG11 medium was inoculated with Synechocystis wild type or mutants harbouring the gfp gene (SNnK.gfp and SNnK.Cgfp mutants' series) to OD730 ≈ 0.1. The cultures were grown to OD730 ≈ 2.5, under continuous light at 30°C, and subsequently diluted to a final OD730 ≈ 0.2 in fresh BG11. Ten millilitres of each diluted culture was transferred to three sterile 25-ml borosilicate glass flasks and closed with cellulose stoppers. The flasks were incubated under the same light and temperature conditions, and samples for total fluorescence measurements were collected after 2, 4, 7, 9 and 11 days after inoculation. For fluorescence measurements, three aliquots of 200 µl of each biological replicate were distributed in a Nunc™ MicroWell™ 96-Well Optical-Bottom Plates (Thermo Fisher Scientific Inc.), and GFP fluorescence and OD790 were detected using the Synergy 2 Multi-Mode Microplate Reader and the Gen5™ software (BioTek Instruments, Inc.). For fluorescence detection, an excitation filter of 485/20 nm and an emission filter of 528/20 nm were used (sensibility set for 110 nm). OD790 was measured using an absorbance filter of 790 nm. Wells with BG11 were included for background fluorescence measurement, and plate readings were carried out in duplicate. Absolute fluorescence values were normalized to the background values of the BG11 medium, and the wild type was used as fluorescence baseline. Mean GFP fluorescence per cell was normalized to the OD790 and is expressed in arbitrary units (A.U.).
2.14. Isobaric tags for relative and absolute quantification
For the proteomics analysis, 50 ml cultures of Synechocystis wild type (WT), the mutants constitutively expressing gfp (SN5K.Cgfp, SN8K.Cgfp, SN10K.Cgfp, SN15K.Cgfp and SN16K.Cgfp) and a mutant carrying a promoterless gfp gene at site N15 (SN15K.gfp) were grown in quadruplicate at 30°C under continuous light, until they reached an OD730 ≈ 0.7. The culture volumes used for sample collection were normalized to the culture with lowest OD730, to guarantee biomass uniformity among samples. The cultures were then centrifuged for 10 min at 3,850 g, and cell pellets were preserved at −80°C prior to protein extraction. The four replicates of each strain were paired and combined together, resulting in 14 samples for analysis (Supplementary Table S5). The samples were run across two isobaric tags for relative and absolute quantifications (iTRAQs) 8-plex with the pooled WT samples being a common control in both experiments creating a point of reference between them.
For protein extraction, cells were washed with 150 mM sodium chloride and spun down for 10 min at 4,000 g, and the supernatant was discarded. Pellets were resuspended in 200 µl of lysis buffer [200 mM triethylammonium bicarbonate (TEAB), 5 mM dithiothreitol (DTT), plant protease inhibitor cocktail and 0.1% (vol/vol) nonidet P40 from Sigma] and transferred to a 2 ml LoBind microcentrifuge tubes (Eppendorf) containing ∼200 mg of 200 µm Zirconium beads. Samples were then pulsed on a bead-beater for 10 cycles of 60 s with a 2-min cooling step on ice between cycles. Samples were centrifuged at 13,000 g for 10 min at 4°C, and supernatants were transferred to new microfuge tubes. Pellets were further resuspended in 200 µl of lysis buffer, pulsed for a further 5 cycles on the bead-beater and centrifuged as before, and supernatants were combined to the previous ones. The combined supernatants were further clarified by centrifugation at 13,000 g for 10 min at 4°C to ensure that any remaining cell debris was removed from the extracts. The clarified extracts (∼400 µl) were incubated with 2 µl of benzonase nuclease (Novagene) for 2 min on ice. Protein content in each extract was precipitated with the addition of four volumes of cold acetone and was incubated overnight at −20°C. The precipitated proteins were then spun down, and the supernatants were discarded. The protein pellets were then resuspended in 200 mM TEAB, and protein concentration was estimated using the modified Lowry spectrophotometric method as described.29 The concentration of the protein extracts and the efficiency of the lysis method were evaluated by sodium dodecyl sulfate polyacrylamide gel.
From each sample, 100 µg of the protein extract was collected for enzymatic digestion, and the final volume was adjusted to 30 µl with 200 mM TEAB, pH 8.5. Proteins were reduced with 10 mM TCEP [tris(2-carboxyethyl)phosphine] for 1 h at 60°C. Cysteines were blocked using 20 mM methyl methanethiosulfonate (MMTS) for 10 min at room temperature. The reduced and blocked proteins were digested overnight at 37°C, using trypsin 1:25 (wt/wt) (Promega) in 200 mM TEAB, pH 8.5. iTRAQ, 8-plex reagents (ABSciex) were resuspended in 100 µl isopropanol and added to the appropriate digested proteins. The peptides and labels were incubated at room temperature for 2 h. After incubation, unreacted iTRAQ reagents were quenched by adding 0.1% (vol/vol) hydroxylamine and incubated for 15 min at room temperature. All labeled peptide fractions were combined and split into two separate tubes. These peptide samples were dried using a vacuum concentrator and stored at −20°C prior to off-line fractionation using a HyperCarb porous graphitic column (particle size: 3 µm, length: 50 mm, diameter: 2.1 mm, pore size: 5 µm, Thermo Scientific) on a UHPLC Ultimate 3000 RS (Dionex). Dried peptides were resuspended in 100 µl of Buffer A [0.1% (vol/vol) trifluoroacetic acid (TFA), 3% (vol/vol) HPLC-grade acetonitrile (ACN) in HPLC-grade water] and were eluted using a linear gradient of Buffer B [0.1% (vol/vol) TFA, 97% (vol/vol) HPLC-grade ACN in HPLC-grade water] ranging from 10 to 60% over 2 h. Peptide elution was monitored using a wavelength of 214 nm and with Chromeleon software, version 6.50 (Dionex). Fractions were collected at 2-min intervals across the gradient and dried using a vacuum concentrator. The dried fractions were stored at −20°C prior to mass spectrometry analysis. The second (reverse-phase) dimension of the chromatographic separation was performed on an Ultimate 3000 capillary HPLC system (Dionex) on-line connected to a quadrupole time-of-flight (Q-ToF) QSTAR XL mass spectrometer (ABSciex). Each HyperCarb fraction was resuspended in loading buffer [0.1% (vol/vol) TFA, 3% (vol/vol) HPLC-grade ACN in HPLC-grade water], and two injections were carried out. Chromatographic separation was performed on a 75 µm × 15 cm C18 analytical column (particle size: 3 µm, pore size: 100 Å, LC Packings, CA, USA) preceded by a C18 trap column (Dionex/LC Packings). A 90-min gradient elution was performed using Buffer A [0.1% (vol/vol) formic acid (FA) and 3% (vol/vol) HPLC-grade ACN in HPLC-grade water] and Buffer B [0.1% (vol/vol) FA and 97% (vol/vol) HPLC-grade ACN in HPLC-grade water], during which Buffer B increased from 3 to 35% at a flow rate of 300 nl min−1. In the mass spectrometer, MS survey scans were acquired at 1 Hz with a m/z (mass-to-charge) range of 350−1,600, and two dynamically selected ions with a +2 or +3 charge state were isolated and fragmented by collision-induced dissociation (CID). The collision energy was optimized to maximize the intensities of the reporter ions without losing sequence information. Each MS/MS (tandem mass spectrum) was accumulated for 3 s. Data were acquired and processed using Analyst QS version 2.0 (ABSciex). Protein identification and quantification were carried out using Phenyx Software version 2.6 (Genebio). MS/MS spectra were searched against the UniProt flat-text database (accessed April 2015), containing 3,576 protein sequences of Synechocystis, with the protein sequences of GFP and NPTII added to it. Mass tolerance was set to 0.3 Da and MS/MS tolerance was set to 0.1 Da. The peptide modifications were the same for each search (fixed modifications: methylthio for cysteines, iTRAQ 8-plex for lysine and peptide N-terminus and variable modifications: oxidation of methionine). The false discovery rate (FDR) was calculated by simultaneously using the true and the reverse databases generated with Phenyx,30 as previously described.31 An FDR of 1% with at least two unique peptides was imposed on the data, and peptide spectral matches (PSMs) falling above this value were excluded. The peptide lists generated by Phenyx were analysed as described elsewhere.32 Each iTRAQ was analysed separately to this point and then merged to generate background proteomic effects across the samples. The peptide quantifications were merged as follows: the lists were checked to find a list of all proteins successfully identified with at least two unique peptides and at least three PSMs in both iTRAQs. The iTRAQ quantification values of these proteins were normalized separately using median correction, to account for internal label variation, before a fixed ‘α’ value (0.5 with this data) was added to all quantifications, to remove the effects of missing labels and noise in the data on the ratio-driven quantification used with iTRAQ. The quantifications were ratio-transformed relative to the mean of the WT controls in each sample and then scaled to each other so that the mean of the WT controls in each sample was equal. A cluster analysis in the form of both a dendrogram (Ward linkage) and principal components analysis (PCA) was then performed to look for background differences between the samples. Significant differences were identified using our in-house protein pipeline, which utilizes a series of paired t-tests with Bonferroni correction, as described elsewhere.33 The free online tool KEGG color-mapper (http://www.genome.jp/kegg/tool/map_pathway2.html, accessed: 27 April 2015) was used to highlight metabolic pathways that were systematically significantly up- or down-regulated.
3. Results
3.1. Selection of putative neutral sites
To identify putative neutral sites in the chromosome of Synechocystis, the list of annotated open reading frames (ORFs) with the respective encoded proteins was retrieved from CyanoBase (http://genome.kazusa.or.jp/cyanobase). From the 3,264 ORFs listed, 1,532 of the putatively encoded proteins were annotated as being unknown or hypothetical, and a subsequent bioinformatics analysis was performed to identify those matching the following criteria: (i) length of the putatively encoded proteins ≤301 amino acids, (ii) encoded proteins with no predicted transmembrane domains, (iii) no reported interaction with other proteins and (iv) no relevant similarities found at the protein sequence level. This analysis led to the selection of 16 candidate neutral sites—termed N1–N16 (Fig. 1 and Supplementary Table S3). Afterwards, a qualitative analysis of the genomic context of these ORFs was performed (Fig. 1B) and revealed that the N2 site superimposes the 5′ region of an ORF (slr0369) putatively encoding a cation or drug efflux system protein, the N4 site overlaps the 5′ region of an ORF (slr0415) putatively encoding a Na+/H+ antiporter and the N9 site corresponds to the ORF located immediately upstream and in the same direction as an ORF (sll0182) putatively encoding an ABC-type transporter. Therefore, these three sites were disregarded, and transcription analyses were performed for the other 13 loci.
3.2. Transcription analysis and final selection
To assess whether there was no transcription or whether the transcript levels of the identified loci were negligible, RT-PCRs were initially performed using RNA extracted from Synechocystis cells collected at the exponential growth phase (OD730 ≈ 0.8). The results revealed high transcript levels for N3, N11, N12 and N13 sites, low transcript levels for N6, N8, N14 and N15 sites, low primer efficiency for N1 site and no detectable amplification for the remaining sites (N5, N7, N10 and N16). Regarding N1 site, the transcription analysis was unreliable, and the locus was too small to design other primers so this site disregarded (Supplementary Fig. S27). Further transcriptional studies were performed collecting cells at different phases (early exponential, exponential and stationary), to evaluate a possible differential transcription of sites N5, N6, N7, N8, N10, N14, N15 and N16 (Fig. 2). The results showed that there was residual transcription for N6 and N14 sites throughout the various growth phases, and therefore, these two sites were disregarded. Concerning the other sites, it was possible to detect residual transcription for N8 and N15 sites, but only at one point of the growth curve—samples collected at the early exponential. Thus, six sites (N5, N7, N8, N10, N15 and N16) were selected to generate mutants to validate their ‘neutrality’. Subsequent cloning difficulties in constructing N7-related plasmids, probably due to secondary structures formation, resulted in this site also being disregarded. The final five target sites (N5, N8, N10, N15 and N16) are evenly distributed throughout the chromosome, with intervals in the order of 60−90° (72° corresponds to 1/5 of the chromosome, see Fig. 1A).
3.3. Generation of mutants
The integrative plasmids used to transform Synechocystis and characterize the neutral sites (Table 1 and Supplementary Table S6) were constructed as described in Material and Methods (for details, see Supplementary Fig. S1). Synechocystis mutants were obtained by natural transformation with the respective plasmid (Table 1 and Supplementary Table S7), and full segregation was confirmed by PCR (Supplementary Fig. S28) and Southern blot (Supplementary Fig. S29). All the mutants produced in this study are fully segregated, with all chromosomal copies mutated and the insertion of ectopic genes was also confirmed (resistance cassettes and gfp modules). The mutants produced using the pSNnKS plasmid series (SNnKS mutants, containing the double selection cassette) exhibited the expected phenotypes of kanamycin resistance (KmR) and sucrose sensitivity (SucS) (Supplementary Fig. S30).
Table 1.
List of organisms and plasmids used/generated in this work
| Organism namea | Description | Source |
|---|---|---|
| Escherichia coli DH5α | Transformation/cloning strain | Invitrogen |
| Synechocystis sp. PCC 6803 | Wild-type strain | Pasteur Culture Collection |
| SNnK | Synechocystis mutants with replacement of the neutral site Nn by a kanamycin resistance cassette | This work |
| SNnKS | Synechocystis mutants with replacement of the neutral site Nn by a kanamycin resistance/sucrose sensitivity cassette | This work |
| SNnK.gfp | Synechocystis mutants with replacement of the neutral site Nn by a kanamycin resistance cassette and the promoterless GFP encoding sequence | This work |
| SNnK.Cgfp | Synechocystis mutants with replacement of the neutral site Nn by a kanamycin resistance cassette and the GFP encoding sequence under the control of a constitutive promoter (PpsbA2*) | This work |
| Plasmid namea | Description | Source |
| pGEM®-T easy | T/A cloning vector | Promega |
| pK18mobsacB | Small mobilizable multi-purpose cloning vector derived from the E. coli plasmids pK18, conferring resistance to kanamycin (nptII), and sensitivity to sucrose (sacB) | National BioResource Project (NIG, Japan): E. coli |
| pSB1A2-E0240 | BioBrick™ vector containing the part BBa_E0240 (GFP generator) | Registry of Standard Biological Parts, MIT |
| pJ201-PpsbA2* | Synthesized minimal psbA2 promoter region, meeting the BioBrick RFC[10]standard specifications, cloned into the delivery vector pJ201 | DNA 2.0 Inc., this work |
| pBSK-FP300 | Synthetic interface containing a BioBrick-compatible multiple cloning site (MunI, NotI, XbaI, SpeI, NotI, PstI) insulated with the transcriptional terminators BBa_B1006 and BBa_B0062 (upstream), and BBa_B0053 and BBa_B020 (downstream) | Epoch Life Science Inc., this work |
| pSNn | pGEM-T easy-based plasmid series containing the BioBrick-compatible interface flanked by the two regions for double homologous recombination on neutral site n | This work |
| pSNnK | pSNn plasmid series with the kanamycin resistance cassette cloned upstream of the BioBrick-compatible interface | This work |
| pSNnKS | pSNn plasmid series with the kanamycin resistance/sucrose sensitivity cassette cloned upstream of the BioBrick-compatible interface | This work |
| pSNnK.gfp | pSNnK plasmid series with the GFP generator BioBrick BBa_E0240 cloned in the BioBrick-compatible interface | This work |
| pSNnK.Cgfp | pSNnK plasmid series with the GFP generator BioBrick BBa_E0240 under the control of a constitutive promoter (PpsbA2*), cloned in the BioBrick-compatible interface | This work |
an = 5, 8, 10, 15, 16.
3.4. Statistical analysis of cyanobacteria-specific growth
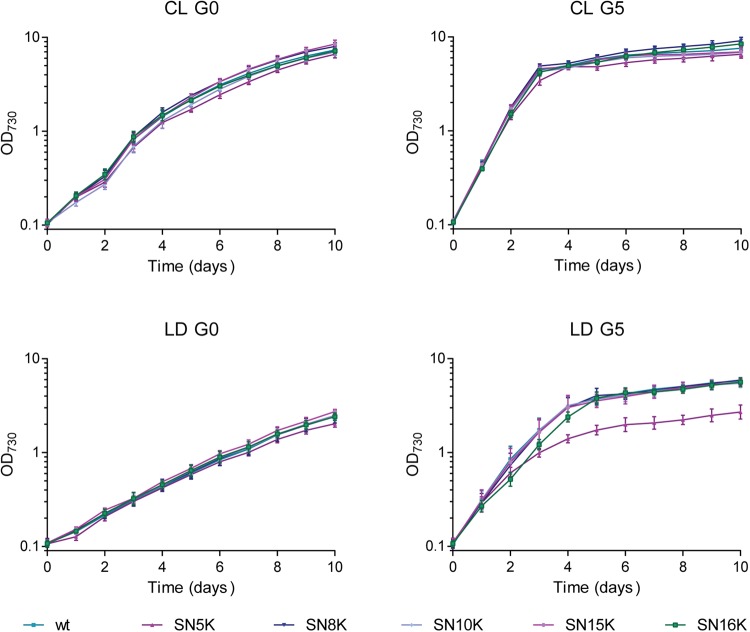
To assess the neutrality of the sites, Synechocystis mutants were produced exchanging each putative neutral site by a kanamycin resistance cassette, using the pSNnK plasmid series (Table 1 and Supplementary Table S6). The successful generation of fully segregated mutants in the five sites showed that the mutations did not affect cell viability under standard growth conditions. The growth of the wild type and the five mutants was evaluated in standard growth conditions for Synechocystis: continuous light (20 µmol photons m−2 s−1; CL) or 12 h light (20 µmol photons m−2 s−1)/12 h dark cycles (LD), in autotrophy (no glucose; G0) or mixotrophy (5 mM glucose; G5), and at a constant temperature of 30°C. The OD of the cultures (OD730) was monitored daily for 10 consecutive days, and the resulting growth curves for each organism and cultivation conditions were obtained (Fig. 3). To assess the effect of the mutations on growth, a statistical analysis of variance (ANOVA) of the growth data was used to compare the wild type and the mutants, using the specific growth rate—µ (see Material and Methods for details). The use of this parameter was considered reliable, since the test duration was the same in all conditions, and growth patterns were similar within the different mutants and replicas. The results of the three-way ANOVA analysis performed on the growth results, including P-values of up to second-order interactions can be found in Supplementary Table S4. Third-order interactions were in accordance to these results. Comparing the effect on growth of each mutation to the wild type, mutants SN5K and SN8K showed highly significant P-values, while for SN10K the P-value is ∼0.01, and for SN15K and SN16K, the P-values are not statistically significant (Supplementary Table S4). In detail, mutant SN5K grows slower than the wild type (particularly in the presence of glucose), while mutant SN8K grows faster under continuous light (with or without glucose). Taking these results into consideration, the chromosomal loci neutrality may be qualitatively ordered as WT ≈ N15 ≈ N16 > N10 > N8 >> N5, in terms of growth. For the remaining variables considered in the ANOVA, irradiance and glucose were the major parameters affecting growth (with highly significant P-values).
Figure 3.
Growth curves of Synechocystis wild type and mutants in the five neutral sites (SN5K, SN8K, SN10K, SN15K, SN16K) cultivated under different conditions. Ten millilitres of culture was grown in 25-ml flasks at 30°C, and OD730 values were recorded daily for 10 consecutive days in continuous light (CL) or 12 h light (20µmolphotons/m2/s)/12 h dark cycles (LD), in autotrophy (G0) or mixotrophy (5 mM glucose, G5). These results are representative of three biological replicates and technical duplicates, and the differences among them are not significant; error bars show ±S.D.
3.5. Synthetic interface robustness and neutral sites functionality
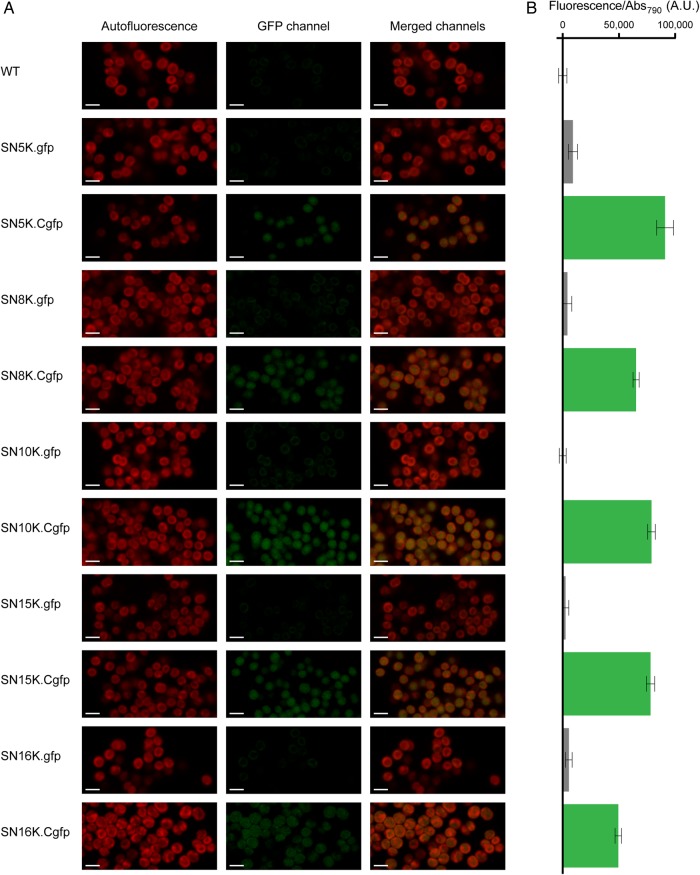
To use the constructed vectors as tools for the integration of synthetic modules, a BioBrick-compatible cloning interface was designed and synthesized. A DNA sequence containing the MunI, NotI, XbaI, SpeI and PstI restriction sites, compatible with the RFC[10] assembly standard, was flanked by two double transcription terminators (BioBricks BBa_B1006 and BBa_B0062 upstream, and BBa_B0053 and BBa_B0020 downstream), to prevent any potential promoter activity from backbone DNA sequences. The interface was synthesized and cloned between the two homologous recombination sequences of each vector. To evaluate the proper transcriptional insulation of the interface, a promoterless green fluorescent protein generator (GFP; BioBrick composite part BBa_E0240) was cloned in the pSNnK plasmid series, and Synechocystis mutants were produced (mutants SNnK.gfp, Table 1 and Supplementary Table S7). Additionally, to assess the functionality of the neutral sites, Synechocystis mutants carrying the GFP generator under the control of a constitutive synthetic promoter (minimal psbA2 promoter, PpsbA2*) were constructed (mutants SNnK.Cgfp, Table 1 and Supplementary Table S7). gfp transcription was assessed by RT-PCR (Supplementary Fig. S31A), and GFP expression was analysed by western blot (Supplementary Fig. S31B), confocal microscopy (Fig. 4A) and fluorimetry (Fig. 4B and Supplementary Fig. S32). In all cases, gfp transcripts and GFP levels could only be detected in mutants where the promoter PpsbA2* is present (SNnK.Cgfp); in these mutants, GFP fluorescence is relatively constant over time (11 days). Moreover, to determine whether the same PpsbA2* promoter-gfp construction is equivalent in terms of transcription strength at the different sites (chromosomal three-dimensional structure can affect promoter strength), RT-qPCR was performed using RNA extracted from the SNnK.Cgfp mutants (wild type was used as negative control). All RT-qPCR parameters in these experiments were in agreement with the MIQE guidelines.28 The normalized fold expression of gfp in the mutants was calculated, and there are no statistically significant differences between the five sites, as calculated by one-way ANOVA (Supplementary Fig. S33).
Figure 4.
Detection of GFP expression in Synechocystis wild type (WT) and insertion mutants containing a promoterless GFP generator (SNnK.gfp) or the GFP generator under the control of the constitutive minimal Ppsba2* synthetic promoter (SNnK.Cgfp). (A) Confocal micrographs of Synechocystis: autofluorescence is depicted in the left column, GFP signal in the middle column and the merged signals in the right column. Scale bars, 2.5 µm. (B) Normalized fluorescence of the cultures analysed in A. Measurements were performed in triplicate, using 200 µl of each culture, and fluorescence was normalized to OD790. Normalized fluorescence from the wild type was used as baseline. Bars indicate mean ± S.D.
3.6. Proteomes of the wild type versus neutral site mutants
To evaluate changes on the proteome of Synechocystis, associated with the deletion of each neutral site/expression of synthetic modules, an iTRAQ analysis was performed. Across the two iTRAQ experiments performed (see Material and Methods for details), 536 of the identified proteins with two or more unique peptides at an FDR of 1% were common to both studies (Supplementary Tables S8–S10). During data analysis, the relative quantifications of the two iTRAQ experiments were scaled, so that the mean of the control samples (wild type) in both iTRAQs was equal. The hierarchical clustering of the iTRAQ labels following combination clearly shows a clustering of the wild-type technical (a and b) and biological (1 and 2) replicates (Supplementary Fig. S34). Close to the wild type, there is a replicate of the mutant containing the promoterless gfp module (SN15K.gfp), while all the mutants constitutively expressing GFP constitute a single cluster (with the biological replicates clustering closely), with the exception of two samples (bSN5K.Cgfp2 and bSN10K.Cgfp2) that do not cluster with any of the others (Supplementary Fig. S34). During data analysis, it became apparent that for these two samples, there was a significantly higher concentration of membrane proteins compared with the other biological replicates from the same set. This was likely due to technical variability during protein extraction and as a result, both the bSN5K.Cgfp2 and bSN10K.Cgfp2 outliers were removed from further analysis. A PCA plot showing the first two components (87.79%, 8.77%) of protein variation (Fig. 5), highlighted that as expected both the heterologous proteins GFP and NPTII (NPTII: aminoglycoside 3′-phosphotransferase, conferring resistance to neomycin/kanamycin) clustered away from the rest of the proteome. Only one native protein, P74485, clustered away with the NPTII protein. Overall, in all cases the proteome was found to be unchanged in any systematic way compared with the wild type, although a reduction of high-abundance phycobiliproteins in the mutants expressing GFP was observed (the effect was not seen in the promoterless gfp module mutant). This reduction was possibly an artefact of the relative quantification methods utilized in the iTRAQ workflow and would require absolute protein quantification against total protein weight per cell to verify.
Figure 5.
PCA of the relative abundances of proteins identified in the iTRAQ experiments for Synechocystis wild type and neutral site mutants containing a gfp module (with or without the constitutive promoter). The first component (x-axis) is responsible for 87.79% of the variation in the samples, and the second component (y-axis) is responsible for 8.79%. The heterologous proteins GFP (P42212) and NPTII (P00552, aminoglycoside 3′-phosphotransferase conferring resistance to neomycin/kanamycin), and a native small stress-related protein (P74485) are clearly separated from the bulk of the proteome (central protein cluster). This figure is available in black and white in print and in colour at DNA Research online.
4. Discussion
Synechocystis is a well-studied cyanobacterium with molecular tools developed for its genetic manipulation. Replicative plasmids have been used to introduce foreign DNA into this organism via conjugation or electroporation.34,35 These plasmids are usually based on the broad-host-range E. coli IncQ plasmid RSF1010 and are not specific for cyanobacteria.9,10 Although presenting some advantages, such as being relatively easy to construct and quick to introduce in the cell, broad-host-range replicative plasmids can exhibit some instability.36,37 In addition, (i) the use of large native DNA fragments may lead to the recombination of the replicative vectors with the chromosome, (ii) removal of selective pressure may lead to loss of the vector and (iii) DNA structure of the vectors may have a strong influence on transcription. Altogether these aspects may compromise the system reliability and behaviour. In addition, it has been shown that Synechocystis' endogenous plasmids copy number per chromosome vary with nutritional conditions and growth phase.38 Taking into consideration that the number of chromosomes also varies throughout growth,39 plasmids copy number calculations may become even more complex. Therefore, stable integration of ectopic DNA into Synechocystis' chromosome is a viable alternative for the implementation of synthetic modules into this photoautotrophic chassis. For this purpose, the availability of fully characterized chromosomal neutral sites is crucial.
To date, only a few chromosomal loci have been used to introduce foreign DNA into Synechocystis, but they have not been extensively characterized. One of these loci disrupts the gene slr016811 that encodes a protein that was shown to be secreted.40 Although its function is unknown, Slr0168 possesses a DUF4114 protein domain, found towards the C-terminal of various bacterial proteins and suggested to carry enzymatic activity.41 An additional locus was described as a ‘nonessential, nontranscribed’12 region; however, RNA-seq works42,43 showed that the genes flanking that insertion locus are transcribed. Another locus described disrupts the ssl0410 gene,14 which is located downstream the gene encoding a subunit of the type I NAD(P)H dehydrogenase complex (ndhB) that is important for CO2 uptake in Synechocystis.44 The intergenic region between slr2030 and slr2031,13 which encodes the regulatory protein RsbU, has also been used for chromosomal integrations. Nonetheless, slr2031 is constitutively transcribed, and the respective mutant showed a defect in growth recovery after nitrogen- and sulfur-starvation-induced stationary phase.45 Furthermore, slr2030 and slr2031 are transcribed as an operon.42,43,45 Consequently, insertions in that intergenic region may disrupt the transcriptional unit and lead to the same phenotype described for slr2031 mutants. More recently, three other neutral sites have been identified,15 based on a RNA-sequencing study,42 and characterized under continuous light, at 30°C. In that study, it was shown that the insertion of a reporter gene in the different sites resulted, however, in different fluorescence intensity. Since the integration sites described previously do not match at least one of the selection criteria established in this study, they were not considered here.
Within this work, several putative neutral sites were identified following an unbiased systematic approach and further characterized for the stable integration of synthetic circuits into Synechocystis' chromosome. Although non-coding DNA (ncDNA) regions could be used for this purpose, they may possess cis-regulatory control sequences, thus affecting the transcription profiles of several genes.46,47 Therefore, in this work, only ORFs putatively encoding unknown or hypothetical proteins were considered. After an initial genomic context analysis of the selected ORFs, RT-PCR transcription studies were performed at different phases of Synechocystis growth curve, and the transcription levels appeared to be negligible for the five sites characterized in this work. These results are partially supported by RNA-sequencing studies,42,43 reporting that the number of direct readings for sites N5, N8 and N16 was close to the cut-off threshold under the tested conditions, while for sites N10 and N15, transcripts were detected mainly in the exponential phase (transcriptional unit TU698),43 or induced by dark (transcriptional unit TU1565),42,43 respectively. This technique allows the high-throughput identification of transcriptional units and may be useful for the identification of additional neutral sites, namely genomic regions that are not transcribed under all the conditions tested. In all cases, complementary validation methods should be used (e.g. RT-qPCR)48 and cell viability and fitness assessed. The successful generation of fully segregated mutants, with each selected neutral site replaced by a kanamycin resistance cassette, showed that they do not encode proteins essential to the viability of the cyanobacterium under the tested growth conditions. Several studies reported that genes crucial to the viability of Synechocystis cannot be successfully and completely inactivated, resulting in heteroploid mutant cells, containing both chromosomes harbouring the wild-type gene and the knockout locus.49,50 Moreover, the neutrality of each site was assessed by analysing the growth of mutants SN5K, SN8K, SN10K, SN15K and SN16K cultivated under different conditions. The results showed that the N15 and N16 sites mutants exhibit growth patterns similar to those of the wild type, while the N5, N8 and N10 site mutants present statistically significant differences. The N5 mutant clearly exhibits a growth decrease, in particular when grown under light/dark cycles in the presence of glucose. Therefore, this should be taken into account when choosing this site for the integration of ectopic DNA. Interestingly, the mutant on site N8 exhibited growth rates slightly higher than the parental strain under continuous light, a feature that may be advantageous for biotechnological applications. As for the remaining variables considered in the ANOVA, and similarly to the results obtained by Lopo and co-workers,51 irradiance and the presence of glucose were the major parameters affecting growth (highly significant P-values).
All the integrative plasmids constructed in this work possess a BioBrick-compatible synthetic interface for cloning purposes. The robustness of this synthetic interface was assessed using the promoterless GFP generator (BioBrick BBa_E0240), which showed no leakiness from the chromosomal backbone for each site, since no/negligible levels of gfp transcripts were detected by RT-PCR, and no GFP expression was observed by western blot, confocal microscopy or whole cell fluorescence measurements. In contrast, using the same techniques, gfp transcripts and GFP signal could be detected in all the mutants generated bearing the GFP generator under the control of a constitutive synthetic promoter (based on the photosensitive promoter of the psbA2 gene of Synechocystis). This shows that these five sites can effectively be used for the functional integration of synthetic modules. Moreover, the relative quantification of transcripts by RT-qPCR showed that gfp transcription is not affected by the position of the neutral site in the chromosome, and consequently, the five sites may be considered equivalent in this respect.
The PCA derived from the iTRAQ experiment showed that the overall proteomes of the neutral site mutants do not change in any systematic way (with the exception of a reduction in the high-abundance phycobiliproteins) compared with the wild type. In addition, the heterologous proteins GFP and NPTII clustered away from the rest of the proteome, an expected result since these proteins are absent in the wild type. Only one native protein, P74485, clustered away from the bulk proteome. This small protein is largely uncharacterized, but has been reported as being related to stress conditions.52,53
Altogether, these results show that the selected chromosomal sites are functional and suitable to receive ectopic DNA. In addition, the vectors constructed in this work followed a rational design that allows the stable integration of synthetic modules into Synechocystis foreseeing, e.g. the production of specialty chemicals. Moreover, the possibility to generate markerless mutants by transforming the SNnKS mutants with the pSNn plamids containing a module of interest enables the use of these vectors to sequentially introduce more than one synthetic module into the chromosome. The existence of more than one integration site, spread throughout the chromosome of Synechocystis, allows the implementation of complex synthetic circuits. The vectors design also considered the possibility to use different resistance cassettes making the implementation faster. This work contributes to the field of synthetic biology by allowing the use of Synechocystis as a photoautotrophic chassis, providing not only standard BioBrick-compatible vectors to integrate synthetic modules into fully characterized chromosomal neutral sites, but also chassis prone to receive a synthetic module without selective markers. Furthermore, our work constitutes a proof of concept for mapping and validating genomic neutral sites that can be applied to other biological systems.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org
Funding
This work was supported by the European Commission through the Seventh Framework Programme, FP7-ENERGY-2012-1-2STAGE-308518 (CyanoFactory), from EU FP6-NEST-2005-Path-SYN project BioModularH2 (contract no. 043340) and from National Funds through Fundação para a Ciência e a Tecnologia (FCT) (grants SFRH/BD/36378/2007 to F.P., SFRH/BPD/64095/2009 to C.C.P., SFRH/BPD/74894/2010 to P.O.). We also acknowledge the Engineering and Physical Sciences Research Council (EPSRC) for funding (EP/E036252/1) and The University of Sheffield for Scholarship funding. Funding to pay the Open Access publication charges for this article was provided by the European Commission through the Seventh Framework Programme, FP7-ENERGY-2012-1-2STAGE-308518 (CyanoFactory).
Supplementary Material
Acknowledgements
The authors are grateful to C. Harley for reading the manuscript, M. Lopo for the preliminary growth experiments, and the ‘National BioResource Project (NIG, Japan): E.coli’ for providing the plasmid pK18mobsacB.
References
- 1.Madigan M.T., Martinko J.M., Stahl D.A., Clark D.P.. 2010, Brock biology of microorganisms. Benjamin Cummings: San Francisco. [Google Scholar]
- 2.Harwood C.R., Pohl S., Smith W., Wipat A.. 2013, Bacillus subtilis: model Gram-positive synthetic biology chassis. In: Colin H., Anil W. (eds), Methods in microbiology. Academic Press: Amsterdam, The Netherlands, pp. 87–117. [Google Scholar]
- 3.Li M., Borodina I.. 2014, Application of synthetic biology for production of chemicals in yeast Saccharomyces cerevisiae, FEMS Yeast Res., 15, 1–12. [DOI] [PubMed] [Google Scholar]
- 4.Berla B.M., Saha R., Immethun C.M., Maranas C.D., Moon T.S., Pakrasi H.B.. 2013, Synthetic biology of cyanobacteria: unique challenges and opportunities, Front. Microbiol., 4, 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Whitton B.A., Potts M.. 2000, Introduction to the cyanobacteria. In: Whitton B., Potts M. (eds), The ecology of cyanobacteria—their diversity in time and space. Kluwer Academic Publishers: Dordrecht, The Netherlands, pp. 1–11. [Google Scholar]
- 6.Montagud A., Navarro E., Fernandez de Cordoba P., Urchueguia J., Patil K.. 2010, Reconstruction and analysis of genome-scale metabolic model of a photosynthetic bacterium, BMC Syst. Biol., 4, 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaneko T., Sato S., Kotani H. et al. . 1996, Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions, DNA Res., 3, 109–36. [DOI] [PubMed] [Google Scholar]
- 8.Koksharova O.A., Wolk C.P.. 2002, Genetic tools for cyanobacteria, Appl. Microbiol. Biotechnol., 58, 123–37. [DOI] [PubMed] [Google Scholar]
- 9.Heidorn T., Camsund D., Huang H.-H. et al. . 2011, Synthetic biology in cyanobacteria: engineering and analyzing novel functions. In: Chris V. (ed.), Methods enzymol. Academic Press: New York, pp. 539–79. [DOI] [PubMed] [Google Scholar]
- 10.Taton A., Unglaub F., Wright N.E. et al. . 2014, Broad-host-range vector system for synthetic biology and biotechnology in cyanobacteria, Nucleic Acids Res., 42, e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams J.G.K. 1988, Construction of specific mutations in photosystem II photosynthetic reaction center by genetic engineering methods in Synechocystis 6803. In: Packer L., Glazer A.N. (eds), Methods enzymol. Academic Press: New York, pp. 766–78. [Google Scholar]
- 12.Burnap R.L., Qian M., Shen J.R., Inoue Y., Sherman L.A.. 1994, Role of disulfide linkage and putative intermolecular binding residues in the stability and binding of the extrinsic manganese-stabilizing protein to the photosystem II reaction center, Biochemistry US, 33, 13712–18. [DOI] [PubMed] [Google Scholar]
- 13.Aoki R., Goto T., Fujita Y.. 2011, A heme oxygenase isoform is essential for aerobic growth in the cyanobacterium Synechocystis sp. PCC 6803: modes of differential operation of two isoforms/enzymes to adapt to low oxygen environments in cyanobacteria, Plant Cell Physiol., 52, 1744–56. [DOI] [PubMed] [Google Scholar]
- 14.Aoki S., Kondo T., Ishiura M.. 1995, Circadian expression of the dnaK gene in the cyanobacterium Synechocystis sp. strain PCC 6803, J. Bacteriol., 177, 5606–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ng A.H., Berla B.M., Pakrasi H.B.. 2015, Fine-tuning of photoautotrophic protein production by combining promoters and neutral sites in the cyanobacterium Synechocystis sp. strain PCC 6803, Appl. Environ. Microbiol., 81, 6857–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clerico E.M., Ditty J.L., Golden S.S.. 2007, Specialized techniques for site-directed mutagenesis in cyanobacteria. In: Rosato E. (ed.), Circadian rhythms. Humana Press: Totowa, pp. 155–71. [DOI] [PubMed] [Google Scholar]
- 17.Trautmann D., Voß B., Wilde A., Al-Babili S., Hess W.R.. 2012, Microevolution in cyanobacteria: re-sequencing a motile substrain of Synechocystis sp. PCC 6803, DNA Res., 19, 435–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanesaki Y., Shiwa Y., Tajima N. et al. . 2012, Identification of substrain-specific mutations by massively parallel whole-genome resequencing of Synechocystis sp. PCC 6803, DNA Res., 19, 67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stanier R.Y., Kunisawa R., Mandel M., Cohenbaz G.. 1971, Purification and properties of unicellular blue-green algae (order Chroococcales), Bacteriol. Rev., 35, 171–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook J., Russell D.W.. 2001, Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press: New York. [Google Scholar]
- 21.Tamagnini P., Troshina O., Oxelfelt F., Salema R., Lindblad P.. 1997, Hydrogenases in Nostoc sp. strain PCC 73102, a strain lacking a bidirectional enzyme, Appl. Environ. Microbiol., 63, 1801–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leitão E., Pereira S., Bondoso J. et al. . 2006, Genes involved in the maturation of hydrogenase(s) in the nonheterocystous cyanobacterium Lyngbya majuscula CCAP 1446/4, Int. J. Hydrogen Energ., 31, 1469–77. [Google Scholar]
- 23.Ferreira D., Pinto F., Moradas-Ferreira P., Mendes M., Tamagnini P.. 2009, Transcription profiles of hydrogenases related genes in the cyanobacterium Lyngbya majuscula CCAP 1446/4, BMC Microbiol., 9, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schäfer A., Tauch A., Jäger W., Kalinowski J., Thierbach G., Pühler A.. 1994, Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum, Gene, 145, 69–73. [DOI] [PubMed] [Google Scholar]
- 25.Eriksson J., Salih G.F., Ghebramedhin H., Jansson C.. 2000, Deletion mutagenesis of the 5′ psbA2 region in Synechocystis 6803: identification of a putative cis element involved in photoregulation, Mol. Cell. Biol. Res. Commun., 3, 292–8. [DOI] [PubMed] [Google Scholar]
- 26.Shibato J.S., Agrawal G.A., Kato H.K., Asayama M.A., Shirai M.S.. 2002, The 5′-upstream cis-acting sequences of a cyanobacterial psbA gene: analysis of their roles in basal, light-dependent and circadian transcription, Mol. Genet. Genomics, 267, 684–94. [DOI] [PubMed] [Google Scholar]
- 27.Pinto F., Pacheco C.C., Ferreira D., Moradas-Ferreira P., Tamagnini P.. 2012, Selection of suitable reference genes for RT-qPCR analyses in cyanobacteria, PLoS One, 7, e34983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bustin S.A., Benes V., Garson J.A. et al. . 2009, The MIQE Guidelines: minimum information for publication of quantitative real-time PCR experiments, Clin. Chem., 55, 611–22. [DOI] [PubMed] [Google Scholar]
- 29.Bensadoun A., Weinstein D.. 1976, Assay of proteins in the presence of interfering materials, Anal. Biochem., 70, 241–50. [DOI] [PubMed] [Google Scholar]
- 30.Reidegeld K.A., Eisenacher M., Kohl M. et al. . 2008, An easy-to-use Decoy Database Builder software tool, implementing different decoy strategies for false discovery rate calculation in automated MS/MS protein identifications, Proteomics, 8, 1129–37. [DOI] [PubMed] [Google Scholar]
- 31.Elias J.E., Gygi S.P.. 2007, Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry, Nat. Methods, 4, 207–14. [DOI] [PubMed] [Google Scholar]
- 32.Ow S.Y., Salim M., Noirel J., Evans C., Rehman I., Wright P.C.. 2009, iTRAQ underestimation in simple and complex mixtures: “the good, the bad and the ugly”, J. Proteome Res., 8, 5347–55. [DOI] [PubMed] [Google Scholar]
- 33.Pham T.K., Roy S., Noirel J., Douglas I., Wright P.C., Stafford G.P.. 2010, A quantitative proteomic analysis of biofilm adaptation by the periodontal pathogen Tannerella forsythia, Proteomics, 10, 3130–41. [DOI] [PubMed] [Google Scholar]
- 34.Matsunaga T., Takeyama H.. 1995, Genetic engineering in marine cyanobacteria, J. Appl. Phycol., 7, 77–84. [Google Scholar]
- 35.Vioque A. 2007, Transformation of cyanobacteria. In: León R., Galván A., Fernández E. (eds), Volume 616, Transgenic Microalgae as Green Cell Factories, Adv. Exp. Med. Biol., Springer: New York, pp. 12–22. [Google Scholar]
- 36.De Gelder L., Ponciano J.M., Joyce P., Top E.M.. 2007, Stability of a promiscuous plasmid in different hosts: no guarantee for a long-term relationship, Microbiology, 153, 452–63. [DOI] [PubMed] [Google Scholar]
- 37.Meyer R. 2009, Replication and conjugative mobilization of broad host-range IncQ plasmids, Plasmid, 62, 57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berla B.M., Pakrasi H.B.. 2012, Up-regulation of plasmid-encoded genes during stationary phase in Synechocystis sp. PCC 6803, a cyanobacterium, Appl. Environ. Microbiol., 78, 5448–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Griese M., Lange C., Soppa J.. 2011, Ploidy in cyanobacteria, FEMS Microbiol. Lett., 323, 124–31. [DOI] [PubMed] [Google Scholar]
- 40.Sergeyenko T.V., Los D.A.. 2000, Identification of secreted proteins of the cyanobacterium Synechocystis sp. strain PCC 6803, FEMS Microbiol. Lett., 193, 213–6. [DOI] [PubMed] [Google Scholar]
- 41.Marchler-Bauer A., Anderson J.B., Chitsaz F. et al. . 2009, CDD: specific functional annotation with the Conserved Domain Database, Nucleic Acids Res., 37, D205–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitschke J., Georg J., Scholz I. et al. . 2011, An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803, Proc. Natl Acad. Sci. USA, 108, 2124–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kopf M., Klähn S., Scholz I., Matthiessen J.K.F., Hess W.R., Voß B.. 2014, Comparative analysis of the primary transcriptome of Synechocystis sp. PCC 6803, DNA Res., 21, 527–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohkawa H., Price G.D., Badger M.R., Ogawa T.. 2000, Mutation of ndh genes leads to inhibition of CO2 uptake rather than HCO3− uptake in Synechocystis sp. strain PCC 6803, J. Bacteriol., 182, 2591–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huckauf J., Nomura C., Forchhammer K., Hagemann M.. 2000, Stress responses of Synechocystis sp. strain PCC 6803 mutants impaired in genes encoding putative alternative sigma factors, Microbiology, 146, 2877–89. [DOI] [PubMed] [Google Scholar]
- 46.Zhou H., Hu H., Lai M.. 2010, Non-coding RNAs and their epigenetic regulatory mechanisms, Biol. Cell, 102, 645–55. [DOI] [PubMed] [Google Scholar]
- 47.Morris K.V. 2012, Non-coding RNAs and the epigenetic regulation of gene expression: drivers of natural selection. Caister Academic Press: Norfolk, UK. [Google Scholar]
- 48.Git A., Dvinge H., Salmon-Divon M. et al. . 2010, Systematic comparison of microarray profiling, real-time PCR, and next-generation sequencing technologies for measuring differential microRNA expression, RNA, 16, 991–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ishii A., Hihara Y.. 2008, An AbrB-like transcriptional regulator, Sll0822, is essential for the activation of nitrogen-regulated genes in Synechocystis sp. PCC 6803, Plant Physiol., 148, 660–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oliveira P., Lindblad P.. 2008, An AbrB-like protein regulates the expression of the bidirectional hydrogenase in Synechocystis sp. strain PCC 6803, J. Bacteriol., 190, 1011–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lopo M., Montagud A., Navarro E. et al. . 2012, Experimental and modeling analysis of Synechocystis sp. PCC 6803 growth, J. Mol. Microbiol. Biotechnol., 22, 71–82. [DOI] [PubMed] [Google Scholar]
- 52.Prakash J.S.S., Sinetova M., Zorina A. et al. . 2009, DNA supercoiling regulates the stress-inducible expression of genes in the cyanobacterium Synechocystis, Mol. Biosyst., 5, 1904–12. [DOI] [PubMed] [Google Scholar]
- 53.Fulda S., Mikkat S., Huang F. et al. . 2006, Proteome analysis of salt stress response in the cyanobacterium Synechocystis sp. strain PCC 6803, Proteomics, 6, 2733–45. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.