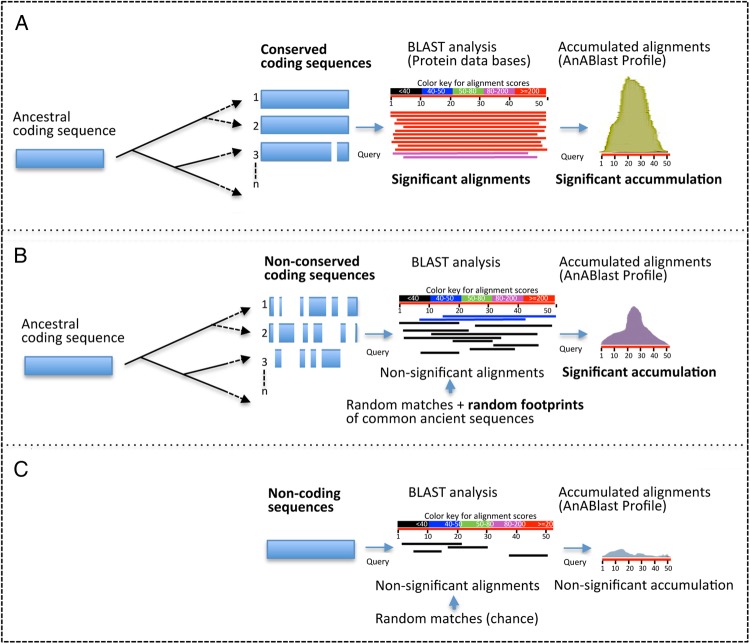
Figure 1.
Schematic representation of the AnABlast strategy for the identification of coding sequences. (A) In conserved proteins, conventional BLAST analysis of query sequences usually generates a number of significant alignments that allow the identification of coding sequences. The accumulation of these alignments along the query sequence (AnABlast profile) generates prominent peaks that also allow the easy identification of conserved coding regions. (B) In non-conserved sequences, BLAST search generates non-significant alignments, but AnABlast profiles highlight coding regions by the significant accumulation of these alignments, generated by random matches as well as from random footprints of common ancestors. (C) Non-coding sequences lack common ancestors in protein databases, and non-significant alignments may only occur by chance. In this case, alignments rarely accumulate in particular regions. This figure is available in black and white in print and in colour at DNA Research online.