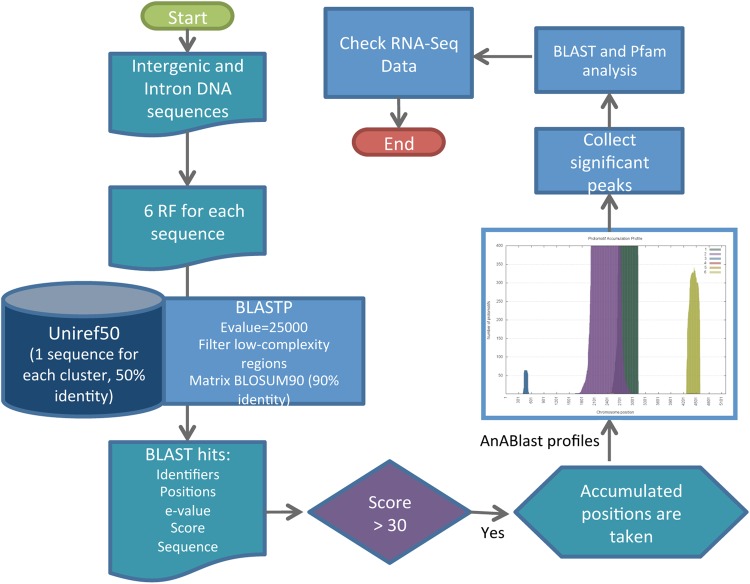
Figure 2.
AnABlast algorithm (schematized flux diagram) for coding sequence identification in a genome-wide search strategy. Amino acid sequences obtained from the six reading frames (RF) of each DNA inter-exon region are subjected to BLAST search against a protein database that minimizes redundancies (uniref50). After low-stringency alignment parameters (optimal bit score threshold: 30), hits are taken at each position of the query amino acid sequence, generating a profile of accumulated alignments along the sequence (AnABlast profile). Significant peaks in the generated profiles (higher than 70 in our S. pombe genome analysis) were selected, and the corresponding amino acid sequence further analysed by conventional BLAST and Pfam search. Ribo-Seq data in the identified genomic region of AnABlast peaks are also analysed. This figure is available in black and white in print and in colour at DNA Research online.