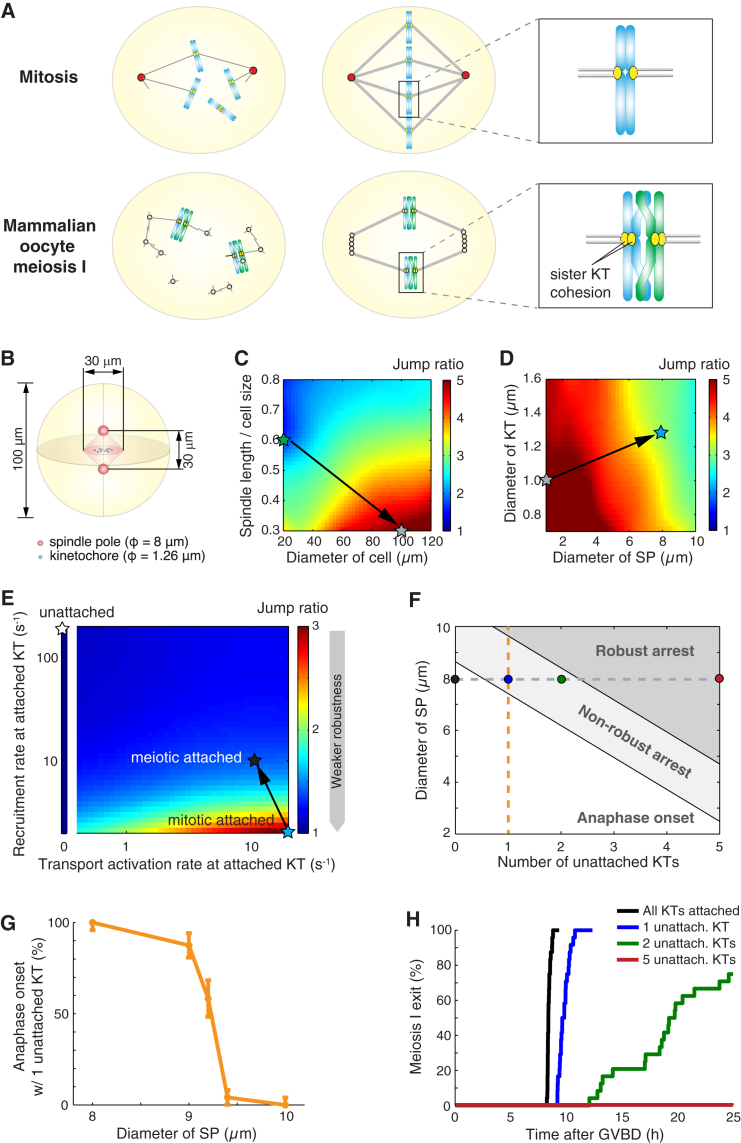
Figure 4.
Nonrobust SAC silencing in oocyte meiosis I cannot result from sizing, but could result from atypical kinetochore attachment. (A) Comparison between somatic cell mitosis and oocyte meiosis I. As opposed to mitosis, oocytes in meiosis I lack centrosomes and maintain unseparated sister kinetochores. (B) Geometric setting for oocyte meiosis I. In (C–E), robustness of the spindle pole signal is characterized by the jump ratio as defined in Fig. 1A. Color-labeled stars represent progressive adjustments of parameters for sizes and rates (details given in the following). (C) Signal robustness at various cell sizes and spindle sizes. (Green star) Sizes for somatic cells. (Gray star) Cell and spindle resized to oocyte dimensions, and the other parameters remain somatic cell values. (D) Signal robustness at various spindle pole sizes and kinetochore sizes. (Gray star) Same as in (C). (Blue star) Cell, spindle, spindle poles, and kinetochores are all resized to oocyte dimensions. (E) Signal robustness under varying recruitment rates and transport activation rates at attached kinetochores. (Blue star) Same as in (D); rates for attached kinetochores follow those in mitosis. (Black star) Increased recruitment rate and decreased transport activation rate at attached kinetochores (see Table S3). (White star) Rates for unattached kinetochores. (F) Meiotic fate with different numbers of unattached kinetochores and at various spindle pole sizes. (White region) Anaphase onset in deterministic simulations. (Light-gray region) Arrest in deterministic simulations but anaphase onset within 25 h in stochastic simulations. (Dark-gray region) Arrest within 25 h in stochastic simulations. (Solid circles) Default parameter value. (Colored dots and orange dashed line) Correspondence to cases in (G) and (H), respectively. (G) Computed frequency of premature anaphase onset with one persistently unattached kinetochore and different spindle pole sizes (parameters along orange dashed line in F). Results from 24 stochastic simulations. Error bars show mean ± SE estimated via the Wald method. (H) Cumulative distribution of time of oocyte meiosis I exit with respect to germinal vesicle breakdown (GVBD) with different numbers of persistently unattached kinetochores. For each simulation, the time for meiotic exit after GVBD = 5 h mean duration for establishing kinetochore attachments after GVBD (15, 66) plus the computed time for 90% cyclin B degradation since the last kinetochore attachment. (Line colors correspond to dot colors in F.) See Appendices D and E in the main text, and Supporting Materials and Methods for detailed procedure of stochastic simulations.