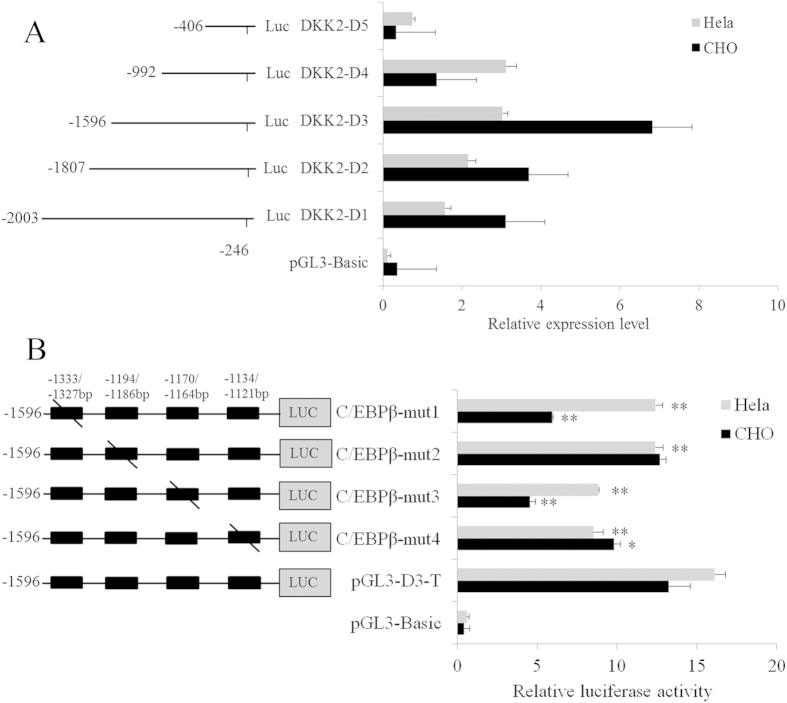
Figure 1. Site-directed mutagenesis and 5′-deletion analysis of C/EBPβ binding sites in the DKK2 promoter.
(A) Promoter activity was analysed in a series of deleted constructs using luciferase assays. Left panel, schematic representation of the mutants linked with the luciferase gene in the pGL3 vector. The nucleotides are numbered from the potential transcription start site, which was assigned +1. Right panel, the relative activity levels of a series of pGL3-D mutant constructs were determined using luciferase assays. (B) Point mutation analysis in the CCAAT boxes of the DKK2 promoter were analysed using luciferase assays. Left panel, schematic structure of site-directed mutagenesis in the putative C/EBPβ binding sites of the DKK2 gene. Right panel, site-directed mutagenesis in the C/EBPβ binding sites of the DKK2 promoter was analysed using luciferase assays. Non-modified pGL3 was used as a negative control. Firefly luciferase activity was normalized to Renilla luciferase activity, and the data are shown as the fold increase/decrease over the luciferase activity observed for pGL3-D3 in CHO and HeLa cells. The data are expressed as the mean ± SE of three replicates. *P < 0.05 and **P < 0.01 compared to the pGL3-D3 construct.