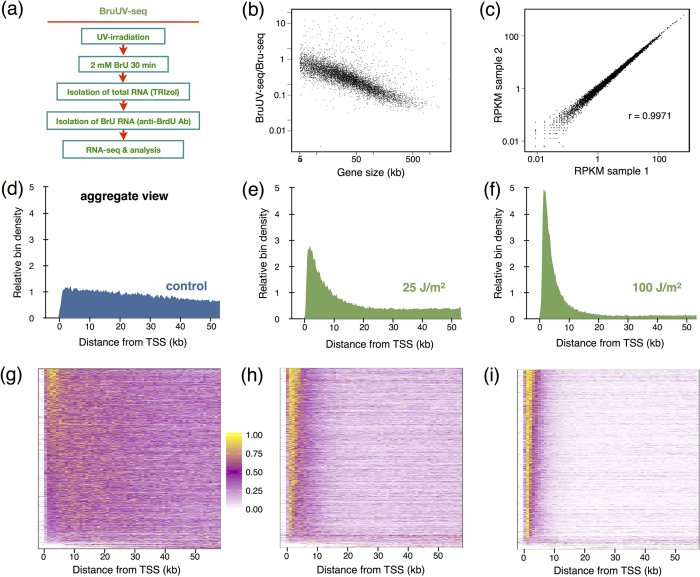
Figure 1.
(a) Experimental outline of the BruUV-Seq technique. (b) Strong correlation between UV-mediated inhibition of transcription and gene size. The ratio of integrated transcription reads over whole genes following UV-irradiation in HF1 human fibroblasts with 20 J/m2 compared to mock-irradiated HF1 cells are shown on the Y-axis while gene size is shown on the X-axis. (c) Reproducibility of BruUV-seq. The RPKM values of the first 5 kb of genes expressed above 0.5 RPKM were compared between two biological experiments involving human HF1 fibroblasts in which cells had been UV-irradiated with 20 J/m2, Bru-labeled and sequenced in parallel. The Pearson’s correlation coefficient between these biological replicates was r = 0.9971. (d) BruUV-seq signal aggregated across genes from K562 cells that had been mock-irradiated, (e), irradiated with UV at 25 J/m2 or (f) 100 J/m2 Genes selected for analysis were at least 50 kb long and expressed more than 1 RPKM in the mock-irradiated sample (1225 genes). Genes were aligned by annotated TSS and median signal was calculated for 250 bp bins and normalized against the mock-irradiated sample. (g–i) Heat maps representing genes (selected as in d) on the vertical axis, and 1 kb bins from −2 kb to + 60 kb on the horizontal axis for (g) mock-irradiated, (h) 25 J/m2 UV, and (i) 100 J/m2 UV. For each gene, bin values were scaled to a 0–1 range and sorted by the sum of scaled values in the first 5 kb following the TSS.