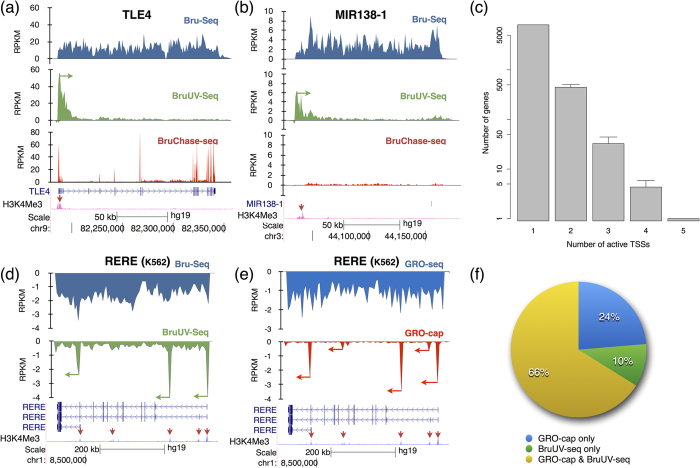
Figure 2. BruUV-seq identifies TSSs genome-wide.
(a) BruUV-seq, using 20 J/m2 UVC, verifies that the TLEA4 gene in HF1 cells is transcribed from a single TSS that aligns with H3K4me3 marks (red arrow). (b) The primary transcript of MIR138–1 is initiating transcription from a single TSS about 80 kb upstream of the MIR138-1 sequence. (c) Genome-wide assessment of the number of TSSs per gene detected with BruUV-seq using HF1 cells and shows that many genes have multiple TSSs. Data is taken from three independent biological experiments with bars showing the standard deviation. (d) BruUV-seq (100 J/m2 UVC) identifies three distinct TSSs in the RERE gene in K562 cells. (e) GRO-cap identifies three strong and two minor TSSs for RERE in K562 cells. (f) Annotated TSSs identified as active in BruUV-seq (100 J/m2 UVC) and GRO-cap. 5720 TSSs represented for genes that expressed at least 1 RPKM in the K562 Bru-seq sample. GRO-cap data was from GEO (GSE60456)18. The H3K4me3 ChIP-seq data are from ENCODE for normal human lung fibroblasts (NHLF) (a,b) and K562 cells (d,e).