Abstract
As an important part of synthetic biology, synthetic promoter has gradually become a hotspot in current biology. The purposes of the present study were to synthesize green tissue-specific promoters and to discover green tissue-specific cis-elements. We first assembled several regulatory sequences related to tissue-specific expression in different combinations, aiming to obtain novel green tissue-specific synthetic promoters. GUS assays of the transgenic plants indicated 5 synthetic promoters showed green tissue-specific expression patterns and different expression efficiencies in various tissues. Subsequently, we scanned and counted the cis-elements in different tissue-specific promoters based on the plant cis-elements database PLACE and the rice cDNA microarray database CREP for green tissue-specific cis-element discovery, resulting in 10 potential cis-elements. The flanking sequence of one potential core element (GEAT) was predicted by bioinformatics. Then, the combination of GEAT and its flanking sequence was functionally identified with synthetic promoter. GUS assays of the transgenic plants proved its green tissue-specificity. Furthermore, the function of GEAT flanking sequence was analyzed in detail with site-directed mutagenesis. Our study provides an example for the synthesis of rice tissue-specific promoters and develops a feasible method for screening and functional identification of tissue-specific cis-elements with their flanking sequences at the genome-wide level in rice.
Synthetic biology, which combines biology and engineering to design and construct new biological parts, devices and systems, has shown great application potentials in many fields1, such as the creation of cells controlled by synthetic genomes2, acquisition of biofuels and pharmaceuticals produced by synthetic gene circuits3, and induction of the precise expression of target genes driven by synthetic promoters4,5. As an important part of synthetic biology, synthetic promoter has gradually become a hotspot in current biology6.
As the most important tool of gene expression regulation in genetic engineering and synthetic biology, promoters can precisely regulate the expression of target genes to expected patterns and levels7,8 and are also crucial for exploring the mechanism of transcriptional regulation7,9,10,11. More and more studies of synthetic promoters have been reported with the development of synthetic biology. Great efforts have been made to construct synthetic promoters in microorganisms. The major strategy is to fuse numerous cis-elements or random sequences with the core promoters and subsequently screen the synthetic promoters whose expression efficiencies are suitable for experimental purposes12,13,14. However, this approach is both labor- and time-consuming for the study of synthetic promoters in plants, especially in crops with long growth periods, such as rice. In animals, recent studies have been focused on combining different promoters to construct synthetic promoter with higher expression efficiency and specificity15,16.
Studies of synthetic promoters in plants are fewer and are mainly based on fusing cis-elements with core promoters. In 2012, Koschmann et al. applied BEST to discover conserved sequence motifs in the promoters of Arabidopsis genes up-regulated by multiple pathogenic stimuli. Then, they compared these motifs with the known cis-regulatory sequences in AthaMap, PLACE and AGRIS databases. The sequences showing no or only low similarity to the already-described regulatory sequences were analyzed with synthetic promoters17. This study provides an enlightening approach for seeking and identifying inducible cis-elements in other species. Another important work was conducted by Liu et al. in 2013, in which pathogen/defense signaling inducible cis-elements were fused with cauliflower mosaic virus (CaMV) 35S/Minimal CaMV 35S to drive a fluorescent protein reporter gene in stable transgenic tobacco and Arabidopsis. The expression of the reporter gene in transgenic plants with bacterial pathogens or phytohormone treatments demonstrated that inducible synthetic promoters can function in transgenic tobacco and Arabidopsis5. A recent progress of synthetic promoters achieved by this team is that they conducted a comprehensive bioinformatic analysis for de novo soybean cyst nematode-inducible motif discovery in the soybean genome and then applied synthetic promoters to identify the candidate motifs in transgenic soybean hairy roots18. This study proves that the new high-throughput screening method has a high application potential in the discovery of inducible cis-elements. Much work in plants has been focused on inducible synthetic promoters, while studies of tissue-specific synthetic promoters have been rarely reported, and there is still no report of any tissue-specific synthetic promoter in rice. The main reason is that few cis-elements with clear functions which are involved in tissue-specific expression have been reported and no effective method for screening these cis-elements has been developed. Therefore, a high-throughput method for screening and identifying cis-elements related to tissue-specific expression is critical to the development of tissue-specific synthetic promoters.
Rice is one of the most important food crops in the world and a model plant for functional genomic researches in cereals19. More complete genomic information20,21,22 and more explicit gene expression information23 greatly facilitate the studies of tissue-specific promoters. Much work has been done to clone tissue-specific promoters and to use them in genetic improvement of rice and gene functional analysis24,25,26,27,28. The above studies lay a solid foundation for seeking tissue-specific cis-elements and constructing tissue-specific synthetic promoters in rice.
In this study, we assembled several short promoters and cis-elements related to tissue-specific expression (PD540-544, POsrbcs-550, POsrbcs-62, EnP3-110, G box and GT1) as well as the first intron of rice Act1 in different combinations, resulting in 5 novel green tissue-specific synthetic promoters which showed different expression efficiencies in various green tissues. Meanwhile, the functions of these expression regulatory sequences in synthetic promoters were also analyzed. As the feasibility of synthesizing tissue-specific promoters in rice was proved, an available method for the discovery of tissue-specific cis-elements is important to the development of tissue-specific synthetic promoters. Subsequently, we scanned and counted the cis-elements in different tissue-specific promoters based on the plant cis-elements database PLACE and the rice cDNA microarray database CREP, resulting in 10 cis-elements whose frequencies in green tissue-specific promoters were relatively higher. Finally, we identified a general regulatory sequence 5′-AAAATATTTAT-3′ (the underlined sequence indicates the core element), which can be applied in the synthesis of green tissue-specific promoters. As flanking sequence may influence the activity of the core element17, we applied site-directed mutagenesis to further analyze the function of the flanking sequence in detail. Our study provides an example for developing tissue-specific synthetic promoters in rice, and proposes a feasible method for screening and functional identification of tissue-specific cis-elements with their flanking sequences at the genome-wide level in rice.
Results
Creation of 5 novel green tissue-specific synthetic promoters (GSSPs)
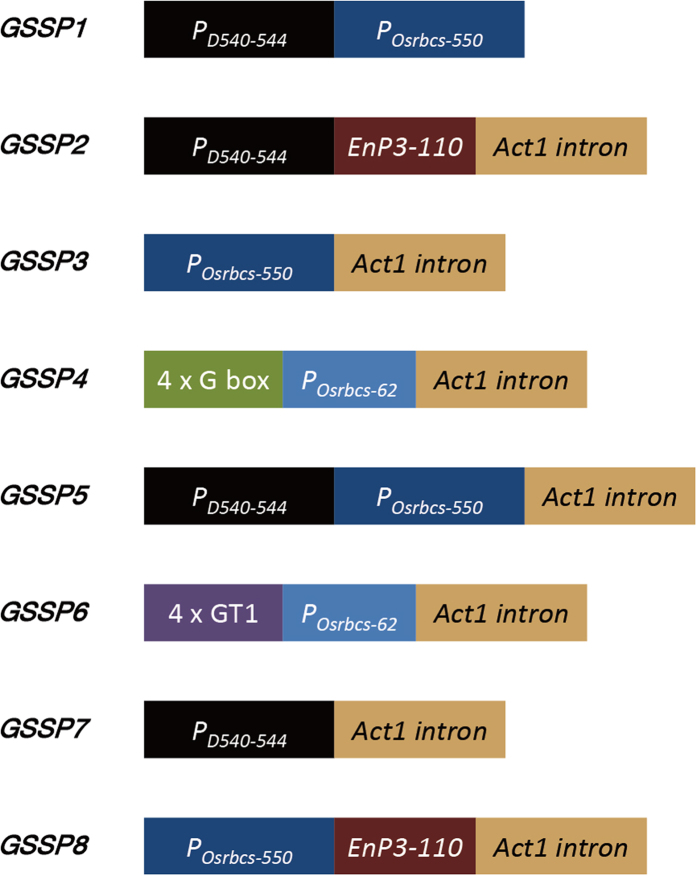
Several regulatory sequences (PD540-544, POsrbcs-550, POsrbcs-62, EnP3-110, the first intron of rice Act1, G box and GT1) (Supplementary Fig. S1) related to tissue-specific expression according to previous reports7,29,30,31,32,33 were used for designing synthetic promoters. As the first intron of rice Act1 is not a promoter but can increase the activity of the promoters adjacent to its upstream29, it was placed in the 3′ region of the synthetic promoters. Considering that the promoter module in the 3′ region of the combinations might be more important to its expression specificity and PD540-544 has activity not only in green tissues but also in root7, PD540-544 was placed away from the 3′ region of the synthetic promoters. If a single cis-element is added in the combinations, it should be quadrupled to increase its effect and placed upstream of a promoter, which is better to be a minimal promoter to avoid the interference from numerous cis-elements5,17. Therefore, G Box or GT1 was quadrupled and placed upstream of POsrbcs-62 minimal promoter as the schemes of GSSP4 or GSSP6 showed (Fig. 1). Meanwhile, the specific combinations were designed for creating new synthetic promoters with desirable expression efficiencies and analyzing the functions of the regulatory sequences in synthetic promoters as well. GSSP1, GSSP3, GSSP5 and GSSP7 were designed to analyze the functions of PD540-544, POsrbcs-550 and the first intron of rice Act1 by comparing the activities of these synthetic promoters simultaneously. Likewise, GSSP2 and GSSP8 were selected to discover the function of EnP3-110 by comparing the activities of GSSP2 and GSSP7 as well as GSSP8 and GSSP3. The schemes of all the synthetic promoters are shown in Fig. 1.
Figure 1. Schemes of 8 synthetic promoters assembled with different expression regulatory sequences.
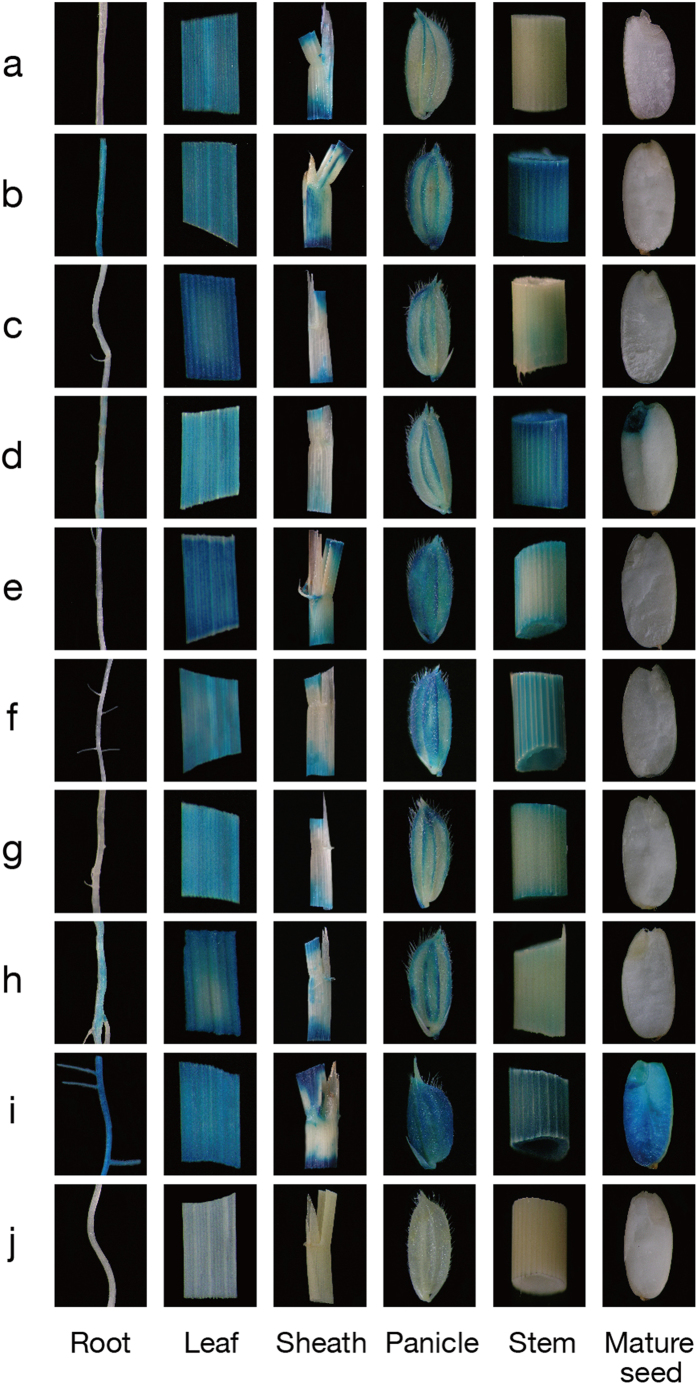
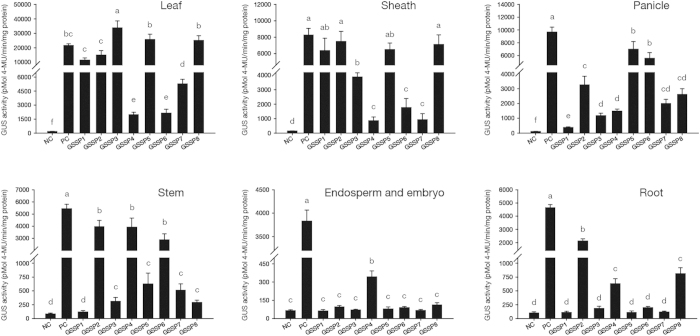
After GUS assays in transgenic plants, the results of histochemical staining in different tissues showed that 5 novel green tissue-specific synthetic promoters (GSSP1, GSSP3, GSSP5, GSSP6, GSSP7) were obtained, which only showed expression efficiencies in all or some of the 4 green tissues: leaf, sheath, panicle and stem (Fig. 2). The GUS fluorometric activities of various tissues in GSSPs transgenic plants (Fig. 3) were in accordance with the histochemical staining results. The expression efficiencies of the 5 synthetic promoters in leaf were ranked as GSSP3 > GSSP5 > GSSP1 > GSSP7 > GSSP6; while in sheath, they were ranked as GSSP1 > GSSP5 > GSSP3 > GSSP6 > GSSP7; in panicle, they were ranked as GSSP5 > GSSP6 > GSSP7 > GSSP3 > GSSP1; in stem, they were ranked as GSSP6 > GSSP5 > GSSP7 > GSSP3 > GSSP1. These synthetic promoters showed different expression efficiencies in various green tissues: GSSP3 and GSSP5 showed higher expression efficiencies than the positive control (CaMV 35S) in leaf, especially GSSP3, whose expression efficiency was 1.5-fold that of the positive control; GSSP1 and GSSP5 showed high expression efficiencies in sheath; while GSSP5 and GSSP6 showed high expression efficiencies in panicle; and only GSSP6 showed high expression efficiency in stem (Fig. 3). Therefore, these green tissue-specific synthetic promoters can be used for realizing efficient expression of different target genes to meet the requirements of various applications. Moreover, they overcome the defects of original regulatory sequences in tissue-specific expression: POsrbcs-62 had no activity in stem32, while GSSP6, in which the GT1 and the first intron of rice Act1 were respectively added to the upstream and downstream of POsrbcs-62, showed sharply increased activity in stem; PD540-544 had activity not only in green tissues but also in root7, while GSSP5, in which POsrbcs-550 and the first intron of rice Act1 were inserted to the downstream of PD540-544, abolished the activity of PD540-544 in root.
Figure 2. Histochemical analysis of GUS expression in various tissues of the transgenic plants containing different synthetic promoters/GUS fusions. (a–h) plants containing GSSP1-GSSP8::GUS; (i) positive control, plants containing CaMV 35S::GUS; (j) negative control, plants containing pDX2181 empty vector.
Figure 3. Quantitative analysis of GUS activity in various tissues of the transgenic plants containing different synthetic promoters/GUS fusions. NC, negative control, plants containing pDX2181 empty vector; PC, positive control, plants containing CaMV 35S::GUS; (a–f ): significant difference (P < 0.05).
Error bars indicate SE based on five independent biological replicates.
Functions of tissue-specific expression related regulatory sequences in synthetic promoters
POsrbcs-550 has been reported to be a truncated green tissue-specific promoter32. According to Fig. 3, by comparing the GUS activity in leaf between GSSP2 and GSSP8 as well as between GSSP3 and GSSP7 transgenic plants, we found that when POsrbcs-550 was used to replace PD540-544, the expression efficiency of the synthetic promoter in leaf was obviously increased. In addition, compared with that of the plants containing GSSP1, the GUS activity of GSSP3, GSSP5 and GSSP8 transgenic plants was more than twice in leaf. Based on the structures of the synthetic promoters, GSSP3, GSSP5 and GSSP8 contained both POsrbcs-550 and the first intron of rice Act1, while GSSP1 did not contain the first intron of rice Act1. Hence, it can be inferred that the coexistence of POsrbcs-550 and the first intron of rice Act1 can sharply increase the activity of synthetic promoters in leaf. According to the GUS activity of GSSP1, GSSP3, GSSP5 and GSSP8 transgenic plants, the synthetic promoters containing POsrbcs-550 showed quite low expression efficiencies in stem. It can be inferred that POsrbcs-550 has suppressive effects on the activity of synthetic promoters in stem, suggesting that POsrbcs-550 contains the cis-elements which inhibit promoter activity in stem.
POsrbcs-62, a further truncated version of POsrbcs-550, had no activity in stem32. However, if it was placed between green tissue-specific cis-elements and the first intron of rice Act1 (GSSP4 and GSSP6), the synthetic promoters showed high expression efficiencies in stem. Compared with the plants containing any synthetic promoters with POsrbcs-550 (GSSP1, GSSP3, GSSP5 and GSSP8), GSSP4 and GSSP6 transgenic plants showed much higher GUS activity in stem. The above results indicate that although POsrbcs-62 had no activity in stem (ie. POsrbcs-62 contained no cis-element which provided promoter activity in stem), the suppressive effect of POsrbcs-550 on promoter activity in stem was also eliminated when being truncated to POsrbcs-62, suggesting that the cis-elements which inhibit promoter activity in stem are present in the region between POsrbcs-550 and POsrbcs-62.
EnP3-110 has been reported to be a short green tissue-specific promoter31. By comparing the GUS activity in GSSP2 and GSSP7 transgenic plants, we found that when EnP3-110 was added in the synthetic promoter, the expression level of the target gene in sheath, stem and root was sharply increased.
PD540-544 was an expression regulatory sequence which had activity in green tissues as well as in root7. GUS activity in the root of GSSP1, GSSP5 and GSSP7 transgenic plants indicated that the activity of PD540-544 in root can be abolished by POsrbcs-550 or the first intron of rice Act1 adjacent to its downstream. The comparison of GUS activity in panicle between GSSP3 and GSSP5 transgenic plants indicated that when PD540-544 was added in the synthetic promoter, the expression level of the target gene in panicle was increased. However, the plants containing GSSP1, which was only composed of PD540-544 and POsrbcs-550, showed quite low GUS activity in panicle. These results indicate that the coexistence of PD540-544 and the first intron of rice Act1 can dramatically increase the activity of synthetic promoters in panicle.
The G Box has been reported as a cis-element related to green tissue expression30. However, by comparing GUS activity in GSSP4 and GSSP6 transgenic plants, we found that when the G Box was placed in the upstream of ‘POsrbcs-62 + the first intron of rice Act1’, activity of the synthetic promoter in non-green tissues embryo and root was observed. If GT1 was used to replace G Box, the expression pattern of the synthetic promoter could be restored to green tissue-specificity and the expression efficiency of the promoter in sheath and panicle was also obviously increased.
The first intron of rice Act1 is an expression regulatory sequence, which can increase the activity of adjacent promoter29. The above results indicate that the coexistence of the first intron of rice Act1 and POsrbcs-550/PD540-544 can greatly increase the activity of the synthetic promoter in leaf/panicle. Meanwhile, analysis of GUS activity in GSSP1 and GSSP5 transgenic plants suggested that the first intron of rice Act1 can not increase the activity of the promoter in sheath, endosperm, embryo and root. These results imply that the increase of expression efficiency by the first intron of rice Act1 shows some tissue-specificity, which might be related to the adjacent promoter34.
Screening of cis-elements involved in green tissue-specific expression
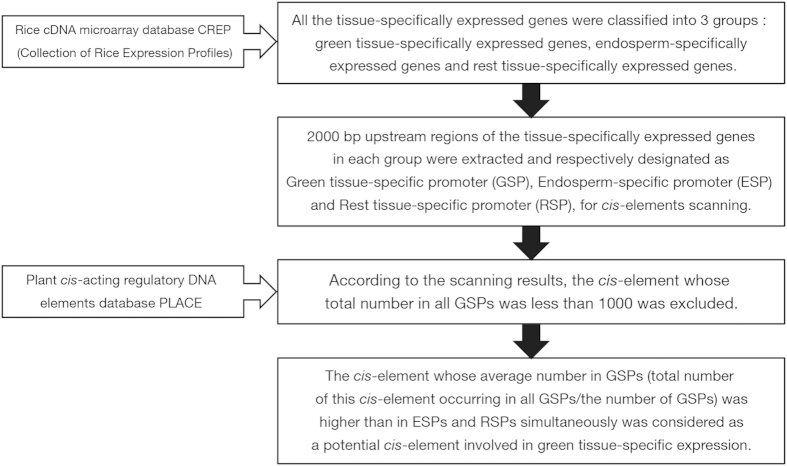
According to the method described in Fig. 4, 10 potential cis-elements involved in green tissue-specific expression were obtained (Table 1). The frequencies and total numbers of these cis-elements in bioinformatic identification are shown in Table 2. Among them, GT1 and GATABOX are known cis-elements for light-regulation and green tissue-specific expression30,33; CACTFTPPCA1 is involved in mesophyll-specific expression35; MYCCONSENSUSAT was reported to be involved in green tissue-specific expression and cold-induction36; and WBOXNTERF3 is related to the activation of gene expression by wounding in leaf37. Besides, RAV1AAT might be related to high expression of transcription factor in leaf and root38. Half of the 10 candidate cis-elements have been reported to be involved in green tissue expression, which proves the availability of our method. In order to find a novel cis-element involved in green tissue-specific expression, core element ROOTMOTIFTAPOX1 (5′-ATATT-3′, here designated as GEAT which stands for Green tissue-specifically Expressed AT-rich element), which is not related to green tissue-specific expression based on the existing annotation, was selected for further study.
Figure 4. Bioinformatic identification of cis-elements involved in green tissue-specific expression.
Table 1. Sequences and annotations of 10 potential green tissue-specific cis-elements.
| Cis-elements | Sequence | Existing Annotation |
|---|---|---|
| GATABOX30 | GATA | Involved in light-regulation and green tissue-specific expression |
| GT133 | GRWAAW | Involved in light-regulated expression |
| CACTFTPPCA135 | YACT | Involved in mesophyll-specific expression |
| MYCCONSENSUSAT36 | CANNTG | Involved in leaf and silique-specific expression and cold-induction |
| WBOXNTERF337 | TGACY | Involved in activation of gene expression by wounding in tobacco leaf |
| ROOTMOTIFTAPOX148 | ATATT | Involved in gene expression in root |
| RAV1AAT38 | CAACA | Binding site of RAV1, which were highly expressed in rosette leaf and root |
| ARR1AT56 | NGATT | Involved in activation of gene expression by the cytokinin-regulated transcription factor |
| WRKY71OS57 | TGAC | Binding site of rice WRKY71, a transcriptional repressor of the gibberellin signaling pathway |
| CURECORECR58 | GTAC | Involved in copper- and oxygen-response |
Cis-elements which have been reported to be involved in green tissue-specific expression are indicated in bold.
Table 2. Average numbers (ANs) in GSPs, ESPs and RSPs and total numbers (TNs) in GSPs of 10 potential green tissue-specific cis-elements.
| Cis-elements |
ANs |
TNs | ||
|---|---|---|---|---|
| GSPs | ESPs | RSPs | ||
| CACTFTPPCA1 | 28.5 | 26.9 | 25.9 | 5980 |
| ARR1AT | 17.9 | 16.3 | 16.5 | 3749 |
| MYCCONSENSUSAT | 16.9 | 15.9 | 15.7 | 3556 |
| GT1 | 14.7 | 13.9 | 13.5 | 3078 |
| GATABOX | 14.6 | 13.9 | 13.3 | 3073 |
| WRKY71OS | 12.7 | 11.7 | 11 | 2658 |
| CURECORECR | 11.3 | 10.6 | 10.2 | 2366 |
| ROOTMOTIFTAPOX1 | 10.9 | 9.1 | 9.3 | 2296 |
| WBOXNTERF3 | 6.4 | 5.3 | 5.0 | 1337 |
| RAV1AAT | 5.3 | 4.9 | 4.8 | 1111 |
(AN of a cis-element in GSPs = TN of this cis-element occurring in all GSPs/the number of GSPs).
Bioinformatic analysis, identification and site-directed mutagenesis of GEAT flanking sequence
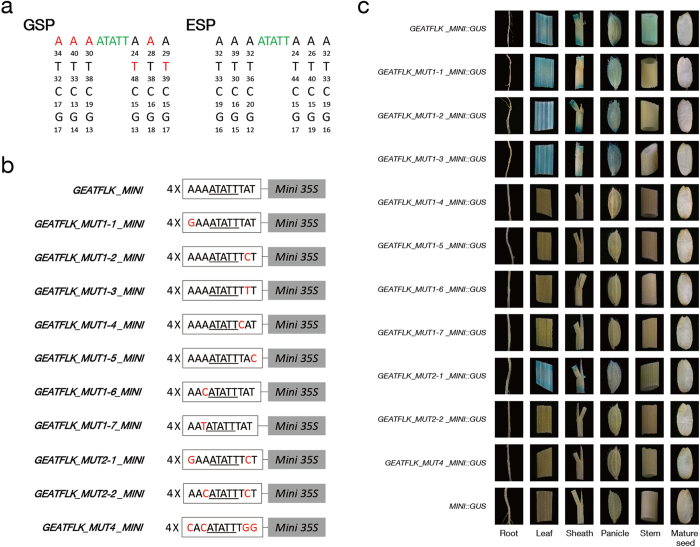
According to the scanning and statistical results of GEAT in GSPs and ESPs (Fig. 5a), GEAT and its flanking sequence (GEATFLK) was determined as 5′-A1A2A3ATATTT4A5T6-3′ (The underlined sequence indicates GEAT). The method for the determination of GEAT flanking sequence was described in Methods. As the presence of TATABOX-like sequence led to the increase of frequency of T at the third site, A was set as the optimal base of the third site in order to avoid interference. Tetramer of GEATFLK was placed upstream of -46 Minimal 35S to promote the expression of GUS (Fig. 5b). The results of GUS assays in transgenic plants showed that the target gene was specifically expressed in leaf, sheath, panicle and stem (Fig. 5c). Thus, it can be confirmed that we successfully identified a novel green tissue-specific cis-element (GEAT) with its flanking sequence.
Figure 5. Bioinformatic and experimental analysis of GEAT flanking sequence.
(a) Frequencies (%) of GEAT flanking sequence bases (3 left and 3 right) in GSPs and ESPs, respectively. Red letters indicate the bases chosen as optimal bases. (b) Schemes of synthetic promoters for functional identification of GEAT with its flanking sequence. Tetramer of GEAT and its flanking sequence (4 × GEATFLK / 4 × GEATFLK_MUT) was placed upstream of −46 Minimal 35S. The underlined sequences indicate the core element GEAT. Mutation sites are indicated by red letters. (c) Histochemical analysis of GUS expression in transgenic plants for functional identification of GEAT with its flanking sequence. MINI::GUS, negative control, plants containing −46 Minimal 35S-pDX2181.
Flanking sequence may influence the activity of core element17. Therefore, the function of GEAT flanking sequence was further analyzed by mutation assays (Fig. 5b). According to the results of GUS assays in single mutation transgenic plants, mutation at any of A3, T4 and T6 could completely abolish the activity of GEAT, indicating that A3, T4 and T6 are critical for the function of GEAT. Mutation at A1 or A5 could not eliminate the activity of GEAT in green tissues except for the stem, in which GEAT activity was lost (Fig. 5c). It can be inferred that A1 and A5 are important for maintaining the activity of GEAT in stem, and mutation at any of them can change the functional pattern of GEAT.
Since GEAT with single mutation at A1 or A5 still had activity in green tissues except for the stem, A1 and A5 were mutated simultaneously to find if double mutation can influence the functions of GEAT in other tissues. The results indicate that double mutation at A1 and A5 of GEAT resulted in the same expression pattern of the target gene with single mutation at A1 or A5. Hence, it can be inferred that multiple mutations at different flanking bases which have similar effects on GEAT can not produce a different functional pattern of GEAT with single mutation at one of these bases. As the mutation at A5 could still maintain the activity of GEAT but the mutation at A3 could not, A3 and A5 were mutated simultaneously to find out whether mutation at A5 can restore the activity abolished by mutation at A3. The result indicated that the activity of GEAT can not be restored by mutation at A5. Therefore, it can be inferred that the mutation at any critical base results in an irreversible abolishment of GEAT activity, which can not be restored by mutation at other bases. Finally, the result of quadruple mutation in GEAT was consistent with our anticipation: when A1, A3, A5 and T6 were mutated simultaneously, the activity of GEAT was completely abolished.
According to the above results, the flanking sequence which supports the activity of GEAT was identified as 5′-AAAATATTTAT-3′ (the dotted bases are critical for maintaining the function of GEAT).
Discussion
In this study, we fused several regulatory sequences related to tissue-specific expression in 8 different combinations. The GUS assays of transgenic plants confirmed that we successfully created 5 green tissue-specific synthetic promoters and proved the feasibility of synthesizing tissue-specific promoters in rice as well. Meanwhile, these novel synthetic promoters can overcome the defects of original regulatory sequences in tissue-specific expression. They also showed different expression efficiencies in various green tissues and thus can meet the requirements of various applications. GSSP3 showed the highest expression efficiency in leaf, which was 1.5-fold that of the positive control. Therefore, it can be applied to transgenic breeding for improving disease/pest resistance in rice leaf28,39, as well as to the studies of the genes related to leaf senescence and other leaf traits40. GSSP5 showed high expression efficiency in leaf, sheath and panicle. Hence, it can be used for efficient expression of the genes related to blast resistance in rice41 as well as for the studies of photosynthesis-related genes42. Although GSSP2 had activity in root, it showed the highest expression efficiency in sheath and stem among all the synthetic promoters and had no activity in endosperm and embryo. Therefore, it can be used for efficient expression of target genes for resistance to pest (such as striped stem borer, brown planthopper and rice plant weevil) and disease (such as rice sheath blight) in rice43,44,45,46, and it is also helpful to the studies of height-related genes47.
Subsequently, the functions of these expression regulatory sequences in synthetic promoters were analyzed. The results are highly valuable for the theoretical and applied research of synthetic promoters. For example, ‘POsrbcs-550 + the first intron of rice Act1’ or ‘PD540-544 + the first intron of rice Act1’ can be added to the target promoter to achieve a great increase of promoter activity in leaf or in panicle, respectively; EnP3-110 can be added to increase promoter activity in sheath, stem and root; and ‘GT1 + POsrbcs-62 + the first intron of rice Act1’ can be used to enhance promoter activity in sheath, stem and panicle. However, some functions of these expression regulatory sequences were different from those in previous reports. For example, POsrbcs-550, EnP3-110 and G Box are green tissue-specific regulatory sequences. However, POsrbcs-550shows suppressive effects on the synthetic promoter activity in stem; EnP3-110 can greatly increase promoter activity in root; and when G Box is placed in the upstream of ‘POsrbcs-62 + the first intron of rice Act1’, it may increase promoter activity in non-green tissues embryo and root. These instances may arise from the interactions of cis-elements and should be explored in future studies.
Cis-element is an essential part of the synthetic promoter. As the feasibility of synthesizing tissue-specific promoters in rice was proved, an available method for the discovery of tissue-specific cis-elements is significant to the development of tissue-specific synthetic promoters. Therefore, another major aim of the present study was to screen and identify cis-elements related to green tissue-specific expression. With the screening method designed in this study, we obtained 10 potential cis-elements involved in green tissue-specific expression based on the information from the rice cDNA microarray database CREP. Half of the 10 candidate cis-elements have been reported to be involved in green tissue expression, which proves the availability of our method. In order to find a novel cis-element involved in green tissue-specific expression, a core element ROOTMOTIFTAPOX1, which is not related to green tissue-specific expression, was chosen and named as GEAT for further study. The results of GUS assays in GEATFLK_MINI::GUS transgenic plants showed that the target gene was specifically expressed in green tissues. This result demonstrates that we successfully identified a novel green tissue-specific expression related cis-element GEAT with its flanking sequence. Moreover, it further proves the reliability of this screening method.
As flanking sequence may influence the activity of core element17, the function of GEAT flanking sequence was analyzed specifically in this work. We found several bases which were indispensable to the whole function or functional pattern of GEAT: A3, T4 and T6 are critical for the whole function of GEAT, and mutation at any of them can completely abolish the activity of GEAT; A1 and A5 are indispensable for the activity of GEAT in stem, and mutation at either of them can change the functional pattern of GEAT. Furthermore, based on the results of double mutation, we found that multiple mutations at different flanking bases which have similar effects on GEAT can not produce a different functional pattern of GEAT with single mutation at one of these bases, and mutation at a critical base will irreversibly abolish the activity of GEAT.
According to the statistical analysis of flanking bases, the frequency of T4 was 44% in ESPs, while it reached up to 48% in GSPs. This result suggests that T4 is important to GEAT, and is even more important to the function of GEAT in GSPs. Mutation assays also proved that it is indispensable for the function of GEAT. T6 showed similar characteristics with T4: its frequency was 33% in ESPs, and was 39% in GSPs. Our results also indicate that when T6 is mutated, GEAT will completely lose its activity. These results verify the reliability of our method for flanking sequence analysis. We did not perform mutation assay at A2 because whatever bases it was mutated to, several additional expression-promoting cis-elements will be formed in the mutant. Based on its frequency (39% in ESPs and 40% in GSPs), we infer that A2 may play a role in the function of GEAT.
The original annotation of GEAT is a cis-element related to gene expression in root48. However, the previous study only predicted its function with bioinformatic analysis, and did not identify this cis-element with transgenic approach. Besides, for the reason that flanking sequence may influence the activity of the core element17, we speculate that even if GEAT can function in root, its activity still needs the support from some specific flanking sequences. In this work, when we mutated A1 or A5, GEAT could not maintain its activity in stem and the expression pattern of the target gene was changed, which also supports our hypothesis.
The traditional experimental approach for seeking and identifying cis-elements is mainly based on electrophoretic mobility shift assay (EMSA). However, either the weak binding capacity of cis-element and TFs or the low content of the target TFs in nuclear proteins may lead to the dissociation of the complex in gel electrophoresis. Besides, even if the TFs-binding activity of a cis-element has been proved, it remains unknown whether its interaction with TFs activates or represses transcription of genes, and whether the interaction functions constitutively or acts in specific tissues and stages. Therefore, it is still necessary to identify the cis-element with transgenic approaches. There are several instances in our study showing that some active cis-elements can not drive the expression of the target gene. For example, both TATABOX and SEF1MOTIF in GEATFLK_MUT1-6_MINI and GEATFLK_MUT1-7_MINI have been proved to possess TFs-binding activity49,50, but even with these cis-elements, GEATFLK_MUT1-6_MINI and GEATFLK_MUT1-7_MINI still had no activity. Meanwhile, when A1 or A5 was mutated, GEAT still showed activity in green tissues except for the stem, suggesting that even if the cis-element has TFs-binding and expression promoting activity, it is still uncertain whether the function pattern of the cis-element is changed. Therefore, EMSA is not sufficient to clarify the functions of cis-elements in gene expression. In this study, we initially applied synthetic promoters for the identification of tissue-specific cis-elements. This approach overcomes the limitation of EMSA in cis-element analysis and has been successfully applied to identify the function patterns of cis-elements combined with different flanking sequences. Overall, we obtained 5 novel green tissue-specific synthetic promoters which can be widely applied in genetic engineering, and provided an example for the synthesis of tissue-specific promoters in rice. We also developed a feasible method for screening and functional identification of tissue-specific cis-elements with their flanking sequences at the genome-wide level in rice.
Methods
Synthetic promoters vector construction
Sequences of PD540-544, POsrbcs-550, POsrbcs-62, EnP3-110, the first intron of rice Act1, G box and GT1 used here were the same as previous reports7,29,30,31,32,33 (see Supplementary Fig. S1). The above regulatory sequences were respectively derived from: LOC_Os08g10020 (PD540-544), LOC_Os12g17600 (POsrbcs-550), LOC_Os12g17600 (POsrbcs-62), LOC_Os03g55734 (EnP3-110) and LOC_Os03g50885 (the first intron of rice Act1). All of the regulatory sequences above were used to synthesize promoters, and their schemes are shown in Fig. 1. These constructs were synthesized by GenScript and ligated into the promoter functional analysis vector pDX2181 after digesting with Hind III and Pst I11.
Agrobacterium-mediated transformation to rice callus
The sequence-confirmed clones were transformed into the Agrobacterium tumefaciens strain EHA105 by electroporation. Subsequently, all the constructs were introduced into Zhonghua11 (Oryza sativa L. ssp. japonica) by Agrobacterium-mediated transformation. pDX2181 (the negative control) and CaMV 35S-pDX2181 (the positive control) were also introduced into Zhonghua11 in the same way. The callus culture and transformation procedures were carried out as previously described51.
Histochemical and fluorometric analysis of GUS activity
Histochemical staining of GUS activity in rice tissues was conducted essentially as described previously52. Various tissues of T0 transgenic-positive transformants (root, leaf, sheath, panicle, stem and mature seed) were incubated in GUS staining solution (50 mM sodium phosphate at pH 7.0, 10 mM Na2-EDTA, 0.1% Triton X-100, 1 mg/mL X-Gluc, 100 μg/ml chloramphenicol, 1 mM potassium ferricyanide, 1 mM potassium ferrocyanide and 20% methanol) at 37 °C for 2–10 h after 15-min vacuum filtration. After GUS staining, the samples were incubated in 70% ethanol to remove chlorophyll and photographs were taken under a dissecting microscope (Leica MZFLIII).
Quantitative analysis of GUS activity was conducted as previously described53. The total protein concentration in the supernatant was quantified using the Bradford assay54. GUS protein in the supernatant was determined fluorometrically with an INFINITE 200 photometer (Tecan Austria Gmbh, Ltd, Grodig, Austria). GUS activity was determined fluorometrically by measuring the amount of 4-methylumbelliferone (Mu) produced under the catalysis of GUS in 1 mg of total protein per minute.
Screening of cis-elements involved in green tissue-specific expression
The information of various tissue-specifically expressed genes in rice was derived from the rice cDNA microarray database CREP (Collection of Rice Expression Profiles, http://crep.ncpgr.cn)23. All the tissue-specifically expressed genes can be divided into 2 groups based on their expression patterns: green tissue-specifically expressed genes (expressed only in green tissues, such as shoot, leaf, sheath, spikelet, panicle (stage 5) and stem) and other tissue-specifically expressed genes. However, although other tissues are not green tissues, part of them are related to green tissues to some extent, such as panicle (stages 1–4) and plumule23. Among the identified tissues in this study, endosperm has no relationship with green tissues and the number of endosperm-specifically expressed genes is sufficient to exclude the influence of the random arrangement of bases in cis-elements analysis. Therefore, we separated endosperm-specifically expressed genes from other tissue-specifically expressed genes as an independent control in order to avoid the interference from numerous non-green tissue-specifically expressed genes. Hence, we divided all the tissue-specifically expressed genes into 3 groups: green tissue-specifically expressed genes (n = 210), endosperm-specifically expressed genes (n = 164) and rest tissue-specifically expressed genes (n = 1019). 2000 bp upstream regions of these genes were extracted and set as promoters for analysis of cis-elements, which were designated as Green tissue-specific promoter (GSP), Endosperm-specific promoter (ESP) and Rest tissue-specific promoter (RSP), respectively. Based on the information of cis-elements in PLACE database55, frequencies of various cis-elements in GSPs, ESPs and RSPs were scanned and subsequently counted. According to the results, the total number of a single cis-element occurring in all GSPs ranged from 1 to 6000. In order to exclude the influence of random events, the cis-element whose total number in all GSPs was less than 1000 was abandoned. Among the rest cis-elements, the one whose average number in GSPs (total number of this cis-element occurring in all GSPs/the number of GSPs) was simultaneously higher than in ESPs and RSPs was considered as a potential cis-element involved in green tissue-specific expression (Fig. 4).
Bioinformatic analysis, identification and site-directed mutagenesis of GEAT flanking sequence
GEAT flanking sequence composed of six ‘optimal bases’ (3 left and 3 right) was determined based on the scanning and statistical results of GEAT in GSPs and ESPs. A GEAT flanking sequence base was set as an optimal base if its frequency at one site in GSPs was higher than at the corresponding site in ESPs and was also higher than 25%.
Tetramer of GEAT and its flanking sequence (4 × GEATFLK) was placed upstream of −46 Minimal 35S to promote the expression of GUS (GEATFLK_MINI::GUS) (Fig. 5b). Under the premise of no formation of additional expression-promoting cis-elements, the flanking sequence of GEAT was treated with single, double and quadruple mutation. Tetramer of GEAT and its mutant flanking sequence was placed upstream of −46 Minimal 35S to promote the expression of GUS (GEATFLK_MUT_MINI::GUS) (Fig. 5b). All the constructs above and −46 Minimal 35S-pDX2181 (MINI::GUS, the negative control) were transformed into Zhonghua11, respectively. Vector construction, callus culture and transformation as well as histochemical staining of GUS activity were performed as described above.
Additional Information
How to cite this article: Wang, R. et al. Novel green tissue-specific synthetic promoters and cis-regulatory elements in rice. Sci. Rep. 5, 18256; doi: 10.1038/srep18256 (2015).
Supplementary Material
Acknowledgments
This research was supported by the National Program of Transgenic Variety Development of China (2014ZX08001001), the National High Technology Research and Development Program of China (863 Program) and the National Natural Science Foundation of China.
Footnotes
Author Contributions Y.L. and R.W. conceived and designed the experiments. R.W. and M.Z. performed the experiments. R.W. performed the data analysis. R.W., Y.L. and R.Y. wrote the paper. Z.L., F.Z. and H.C. revised the paper. Y.L. secured the funds to support this research.
References
- Baltes N. J. & Voytas D. F. Enabling plant synthetic biology through genome engineering. Trends Biotechnol. 33, 120–131 (2014). [DOI] [PubMed] [Google Scholar]
- Gibson D. G. et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science 329, 52–56 (2010). [DOI] [PubMed] [Google Scholar]
- Slusarczyk A. L., Lin A. & Weiss R. Foundations for the design and implementation of synthetic genetic circuits. Nat. Rev. Genet. 13, 406–420 (2012). [DOI] [PubMed] [Google Scholar]
- Chen Y. S. et al. A late embryogenesis abundant protein HVA1 regulated by an inducible promoter enhances root growth and abiotic stress tolerance in rice without yield penalty. Plant Biotechnol. J. 13, 105–116 (2015). [DOI] [PubMed] [Google Scholar]
- Liu W. et al. Bacterial pathogen phytosensing in transgenic tobacco and Arabidopsis plants. Plant Biotechnol. J. 11, 43–52 (2013). [DOI] [PubMed] [Google Scholar]
- Venter M. Synthetic promoters: genetic control through cis engineering. Trends Plant Sci. 12, 118–124 (2007). [DOI] [PubMed] [Google Scholar]
- Cai M., Wei J., Li X., Xu C. & Wang S. A rice promoter containing both novel positive and negative cis-elements for regulation of green tissue-specific gene expression in transgenic plants. Plant Biotechnol. J. 5, 664–674 (2007). [DOI] [PubMed] [Google Scholar]
- Yi N. et al. Analysis of the Wsi18, a stress-inducible promoter that is active in the whole grain of transgenic rice. Transgenic Res. 20, 153–163 (2011). [DOI] [PubMed] [Google Scholar]
- Balasubramani A. et al. Deletion of a Conserved cis-Element in the Ifng Locus Highlights the Role of Acute Histone Acetylation in Modulating Inducible Gene Transcription. PLoS Genet. 10, e1003969 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walcher C. L. & Nemhauser J. L. Bipartite promoter element required for auxin response. Plant Physiol. 158, 273–282 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye R., Zhou F. & Lin Y. Two novel positive cis-regulatory elements involved in green tissue-specific promoter activity in rice (Oryza sativa L ssp.). Plant Cell Rep. 31, 1159–1172 (2012). [DOI] [PubMed] [Google Scholar]
- Rytter J. V. et al. Synthetic promoter libraries for Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 98, 2617–2623 (2014). [DOI] [PubMed] [Google Scholar]
- Sohoni S. V., Fazio A., Workman C. T., Mijakovic I. & Lantz A. E. Synthetic Promoter Library for modulation of actinorhodin production in Streptomyces coelicolor A3 (2). PloS one 9, e99701 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim S. S., An S. J., Kang M., Lee J. & Jeong K. J. Isolation of fully synthetic promoters for high-level gene expression in Corynebacterium glutamicum. Biotechnol. Bioeng. 110, 2959–2969 (2013). [DOI] [PubMed] [Google Scholar]
- Dai J. et al. The combination of a synthetic promoter and a CMV promoter improves foreign gene expression efficiency in myocytes. J. Biotechnol. 158, 91–96 (2012). [DOI] [PubMed] [Google Scholar]
- Kang W. et al. A macrophage-specific synthetic promoter for therapeutic application of adiponectin. Gene Ther. 21, 353–362 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koschmann J. et al. Integration of bioinformatics and synthetic promoters leads to the discovery of novel elicitor-responsive cis-regulatory sequences in Arabidopsis. Plant Physiol. 160, 178–191 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W. et al. Computational discovery of soybean promoter cis-regulatory elements for the construction of soybean cyst nematode-inducible synthetic promoters. Plant Biotechnol. J. 12, 1015–1026 (2014). [DOI] [PubMed] [Google Scholar]
- Zhang Q. Strategies for developing green super rice. Proc. Natl Acad. Sci. USA 104, 16402–16409 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff S. A. et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296, 92–100 (2002). [DOI] [PubMed] [Google Scholar]
- Yu J. et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296, 79–92 (2002). [DOI] [PubMed] [Google Scholar]
- Pan Y. et al. Comparative BAC-based physical mapping of Oryza sativa ssp. indica var. 93-11 and evaluation of the two rice reference sequence assemblies. Plant J. 77, 795–805 (2014). [DOI] [PubMed] [Google Scholar]
- Wang L. et al. A dynamic gene expression atlas covering the entire life cycle of rice. Plant J. 61, 752–766 (2010). [DOI] [PubMed] [Google Scholar]
- Azria D. & Bhalla P. L. Agrobacterium-mediated transformation of Australian rice varieties and promoter analysis of major pollen allergen gene, Ory s 1. Plant cell rep. 30, 1673–1681 (2011). [DOI] [PubMed] [Google Scholar]
- Ha S. H. et al. Application of two bicistronic systems involving 2A and IRES sequences to the biosynthesis of carotenoids in rice endosperm. Plant Biotechnol. J. 8, 928–938 (2010). [DOI] [PubMed] [Google Scholar]
- Jeong J. S. et al. OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field. Plant Biotechnol. J. 11, 101–114 (2013). [DOI] [PubMed] [Google Scholar]
- Molla K. A. et al. Rice oxalate oxidase gene driven by green tissue-specific promoter increases tolerance to sheath blight pathogen (Rhizoctonia solani) in transgenic rice. Mol. Plant Pathol. 14, 910–922 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye R. et al. Development of insect-resistant transgenic rice with Cry1C*-free endosperm. Pest Manag. Sci. 65, 1015–1020 (2009). [DOI] [PubMed] [Google Scholar]
- McElroy D., Zhang W., Cao J. & Wu R. Isolation of an efficient actin promoter for use in rice transformation. Plant cell 2, 163–171 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald R. & Cashmore A. R. Mutation of either G box or I box sequences profoundly affects expression from the Arabidopsis rbcS-1A promoter. EMBO J. 9, 1717–1726 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye R. Isolation, functional characterization and application of the tissue specific expression promoters from rice. Ph.D. Dissertation, Huazhong Agricultural University, (2012).
- Huang H. & Lin Y. Cloning and functional analysis of the rice rbcS gene promoter. J. Huazhong Agr. Univ. 15, 451–458 (2007). [Google Scholar]
- Lam E. & Chua N. H. GT-1 binding site confers light responsive expression in transgenic tobacco. Science 248, 471–474 (1990). [DOI] [PubMed] [Google Scholar]
- Oszvald M. et al. Development and characterization of a chimaeric tissue-specific promoter in wheat and rice endosperm. In Vitro Cell. Dev. Biol. Plant 44, 1–7 (2008). [Google Scholar]
- Gowik U. et al. Cis-Regulatory Elements for Mesophyll-Specific Gene Expression in the C4 Plant Flaveria trinervia, the Promoter of the C4 Phosphoenolpyruvate Carboxylase Gene. Plant cell 16, 1077–1090 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann U., Sagasser M., Mehrtens F., Stracke R. & Weisshaar B. Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP, and BHLH factors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Mol. Biol. 57, 155–171 (2005). [DOI] [PubMed] [Google Scholar]
- Nishiuchi T., Shinshi H. & Suzuki K. Rapid and Transient Activation of Transcription of the ERF3 Gene by Wounding in Tobacco Leaves. J. Biol. Chem. 279, 55355–55361 (2004). [DOI] [PubMed] [Google Scholar]
- Kagaya Y., Ohmiya K. & Hattori T. RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res. 27, 470–478 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F. et al. Plasma membrane localization and potential endocytosis of constitutively expressed XA21 proteins in transgenic rice. Mol. Plant 3, 917–926 (2010). [DOI] [PubMed] [Google Scholar]
- Zhou Y. et al. Identification and functional characterization of a rice NAC gene involved in the regulation of leaf senescence. BMC Plant Biol. 13, 132 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua L. et al. The isolation of Pi1, an allele at the Pik locus which confers broad spectrum resistance to rice blast. Theor. Appl. Genet. 125, 1047–1055 (2012). [DOI] [PubMed] [Google Scholar]
- Li C. et al. A rice plastidial nucleotide sugar epimerase is involved in galactolipid biosynthesis and improves photosynthetic efficiency. PLoS Genet. 7, e1002196 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du B. et al. Identification and characterization of Bph14, a gene conferring resistance to brown planthopper in rice. Proc. Natl Acad. Sci. USA 106, 22163–22168 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J. M., Raghupathy V. & Veluthambi K. Enhanced sheath blight resistance in transgenic rice expressing an endochitinase gene from Trichoderma virens. Biotechnol. Lett. 31, 239–244 (2009). [DOI] [PubMed] [Google Scholar]
- Wang R. et al. Functional analysis of OsPGIP1 in rice sheath blight resistance. Plant Mol. Biol. 87, 181–191 (2015). [DOI] [PubMed] [Google Scholar]
- Yang Z., Chen H., Tang W., Hua H. & Lin Y. Development and characterisation of transgenic rice expressing two Bacillus thuringiensis genes. Pest Manag. Sci. 67, 414–422 (2011). [DOI] [PubMed] [Google Scholar]
- Zhu L. et al. Identification and characterization of SHORTENED UPPERMOST INTERNODE 1, a gene negatively regulating uppermost internode elongation in rice. Plant Mol. Biol. 77, 475–487 (2011). [DOI] [PubMed] [Google Scholar]
- Elmayan T. & Tepfer M. Evaluation in tobacco of the organ specificity and strength of the rolD promoter, domain A of the 35S promoter and the 35S2 promoter. Transgenic Res. 4, 388–396 (1995). [DOI] [PubMed] [Google Scholar]
- Grace M. L., Chandrasekharan M. B., Hall T. C. & Crowe A. J. Sequence and spacing of TATA box elements are critical for accurate initiation from the β-phaseolin promoter. J. Biol. Chem. 279, 8102–8110 (2004). [DOI] [PubMed] [Google Scholar]
- Lessard P. A. et al. Multiple nuclear factors interact with upstream sequences of differentially regulated β-conglycinin genes. Plant Mol. Biol. 16, 397–413 (1991). [DOI] [PubMed] [Google Scholar]
- Hiei Y., Ohta S., Komari T. & Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282 (1994). [DOI] [PubMed] [Google Scholar]
- Jefferson R. A., Kavanagh T. A. & Bevan M. W. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L. et al. Isolation of the endosperm-specific LPAAT gene promoter from coconut (Cocos nucifera L.) and its functional analysis in transgenic rice plants. Plant Cell Rep. 29, 1061–1068 (2010). [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 (1976). [DOI] [PubMed] [Google Scholar]
- Higo K., Ugawa Y., Iwamoto M. & Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27, 297–300 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross E. J. et al. Activation of the Oryza sativa non-symbiotic haemoglobin-2 promoter by the cytokinin-regulated transcription factor, ARR1. J. Exp. Bot. 55, 1721–1731 (2004). [DOI] [PubMed] [Google Scholar]
- Zhang Z. L. et al. A rice WRKY gene encodes a transcriptional repressor of the gibberellin signaling pathway in aleurone cells. Plant Physiol. 134, 1500–1513 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropat J. et al. A regulator of nutritional copper signaling in Chlamydomonas is an SBP domain protein that recognizes the GTAC core of copper response element. Proc. Natl Acad. Sci. USA 102, 18730–18735 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.