Figure 2. ChIP-QPCR for histone modifications (H3K4me1 and H3K27ac) in human liver cancer cell (LC control) and normal liver cell (CC control).
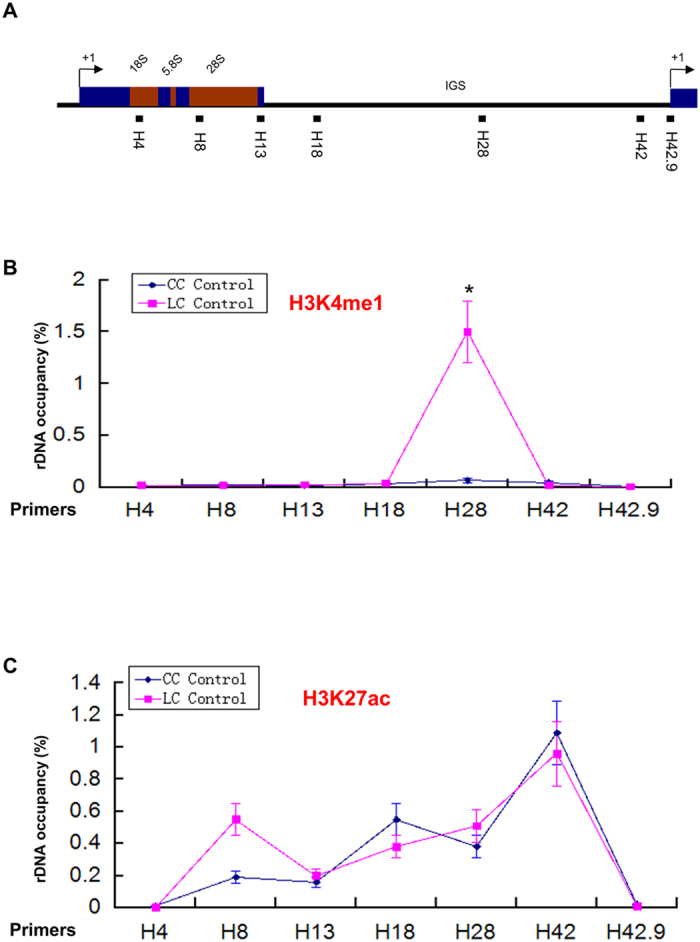
(A) Schematic representation of one human rDNA repeat. The positions of QPCR amplicons in ChIP assays are indicated with solid bars. (B) Enrichment of H3K4me1 on rDNA analyzed by ChIP-QPCR. Chromatin from HepG2 cells was cross-linked and immunoprecipitated with H3K4me1 antibody, DNA was analyzed by QPCR with different sets of primers indicated in (A). The percentage of DNA associated with anti-H3K4me1 antibody was calculated relative to the DNA from ChIP input. Values are represented by means ± SD derived from three independent ChIP experiments, each experiment was tested by at least three independent QPCR reactions. Student’s t-test was performed between human liver cancer cell (LC Control) and normal liver cell (CC Control). (C) Enrichment of H3K27ac on rDNA analyzed by ChIP-QPCR. Chromatin from HepG2 cells was cross-linked and immunoprecipitated with H3K27ac antibody, DNA was analyzed by QPCR with different sets of primers indicated in (A). The percentage of DNA associated with anti-H3K27ac antibody was calculated relative to the DNA from ChIP input. Values are represented by means ± SD from three independent ChIP experiments, each experiment was tested by at least three independent QPCR reactions. Student’s t-test was performed between human liver cancer cell (LC Control) and normal liver cell (CC Control).
