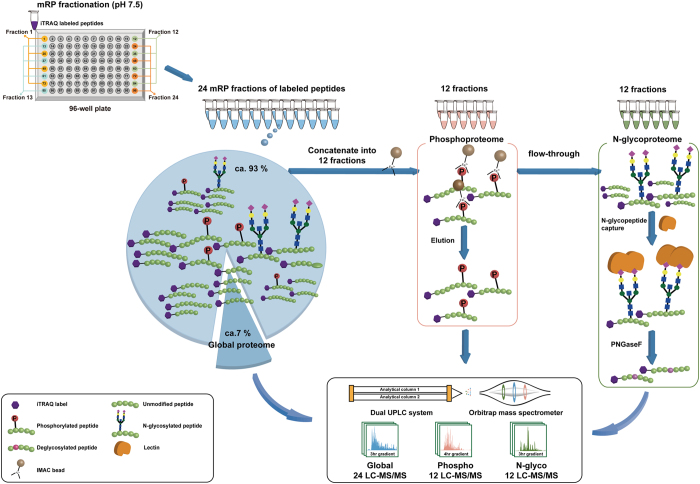
Figure 1. Overall workflow in the SEPG method for integrated profiling of global proteome, phosphoproteome, and N-glycoproteome.
First, iTRAQ-labeled peptides were fractionated into 24 fractions using mRP fractionation on the 96-well plate as schematically described (top left). For each of 24 fractions, global profiling was then performed, and the remaining 24 fractions were further concatenated into 12 fractions. For each of the 12 fractions, IMAC enrichment was performed, followed by phosphoproteome profiling. Finally, for the flow-throughs of the 12 fractions from the IMAC experiments, N-glycopeptide enrichment was performed, followed by N-glycoproteome profiling.