Fig. 1.
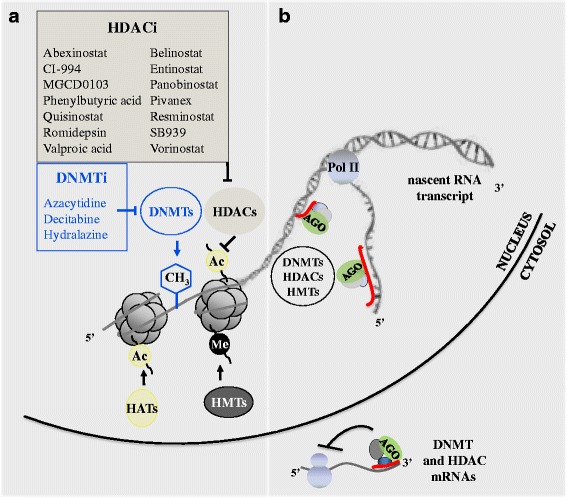
Schematic representation of gene expression regulation by epigenetic drugs, components of the DNA and chromatin-modifying machinery and ncRNAs. a Epigenetic drugs reported to be effective against cancer cells inhibit the activity of DNA methyltransferases (DNMTi) or histone deacetylases (HDACi). DNMTs add a methyl group (CH3) to the 5′ carbon atom of cytosine in DNA CpG dinucleotides. DNMTs also participate in multiprotein chromatin-modifying complexes containing histone deacetylases (HDACs) and histone methyltransferases (HMTs),which induce post-translational modifications of lysine residues in the amino terminal tails of nucleosomal histones, including deacetylation (HDACs), methylation (HMTs) and acetylation (histone acetyltransferases (HAT). Specific molecular modifications on CpGs and nucleosomal histones affect the higher order of chromatin architecture and function by changing the interaction of histones with DNA or the contact between different histones in adjacent nucleosomes. This allows or denies the accessibility of the transcriptional machinery and DNA-binding proteins to specific sites on genome, resulting in activation or silencing of gene transcription. Ac acetylation, Me methylation. b Short and long ncRNA are emerging as novel regulators of chromatin structure, alternative to DNA-binding proteins. They can act as key specificity determinants for epigenetic regulation of gene expression. In the nucleus, both short and long ncRNAs can bind complementary sequences on DNA or nascent RNA transcripts and guide the Argonaute-containing complexes (Ago) to recruit HDACs, HMTs and DNMTs for gene silencing. Nascent lncRNAs can also be tethered to the locus from which they are transcribed through association with RNA polymerase II (Pol II). In the cytosol, microRNAs and siRNAs act as post-transcriptional regulators of the expression of HDAC and DNMTs through their complementarity with mRNA sequences
