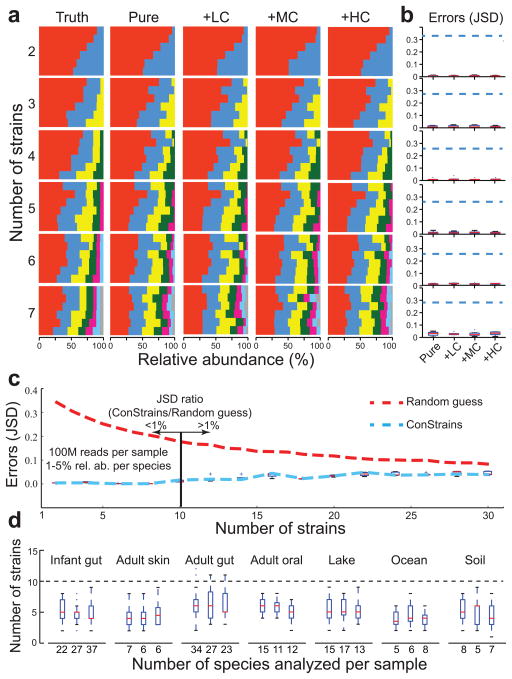
Figure 2. ConStrains correctly predicts the strain composition of in silico-simulated data sets.
A comparison of true and predicted strain composition profiles of in silico-simulated multi-strain mixtures is shown. (a) An increasing number of multi-strain mixtures (n = 2–7; rows) was analyzed with ConStrains either containing only the target strains (pure) or in the context of a metagenome of low, medium, and high complexity (+LC, +MC, and +HC, respectively). In each box of barcharts, the colors represent different strains that were mixed in six different ratios (x axis, relative abundance) with a Shannon index (y axis) increasing from top to bottom. In the resulting 144 admixtures, all strains were correctly identified. (b) To compare the predictions in abundance for each strain, the Jensen-Shannon Divergence (JSD) between predicted composition and the true composition was determined. Blue dashed lines mark the expected errors from random guesses. The box marks the interquartile range, the red bar marks the interquartile median, whiskers represent the top and the bottom 25% data range, and outliers are marked by crosses. Good performance was obtained for all compositions, with minimal difference in the accuracy of results between pure mixtures and metagenomic mixtures; see also Supplementary Fig. 3b for more detailed graphs. (c) Graph showing ConStrains’ ability to correctly infer intra-specific structure as a function of the number of strains contained in a sample. Shown is a typical case with the species’ relative abundance ranging from 1% to 5% and a sequencing depth of 100 million paired-end reads, though higher abundance or sequencing depth would improve its accuracy. The ConStrains’ prediction JSD errors (blue dashed line and boxes) were below 1% of null informative prediction errors (random guess; red dashed line) when the number of strains within a species was less than ten. (d) For comparison, three metagenomic samples were randomly chosen from seven different niches, ranging from adult gut microbiome to a marine planktonic community. More than 95% of the species from these metagenomic samples possessed fewer than ten strains (dashed horizontal line). Dashed lines and whiskers mark the interquartile range; plusses mark the outliers.