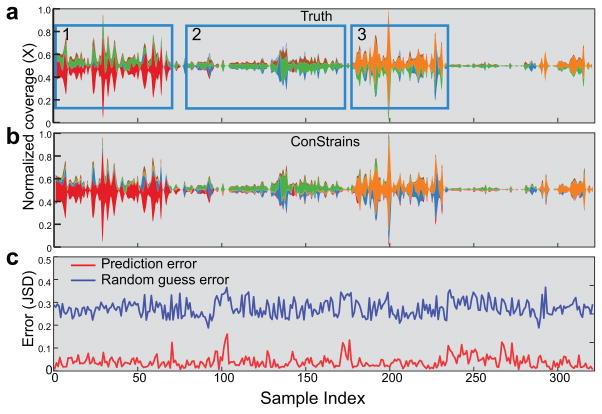
Figure 3.
ConStrains scales to large time series and accurately predicts strain dynamics. In the absence of existing large time series metagenomic data sets, a simulated set with 322 samples was created. Shown are the strain predictions within the Bacteroides fragilis species. The (a) true and (b) ConStrains-predicted relative abundance (y axis) of B. fragilis strains (stream ribbon width, with different colors representing different strains) in different samples sorted in longitudinal order (x axis, sample index) are illustrated. Inset windows 1–3 in a indicate periods with different dominant strains. (c) Prediction errors (red line) in each sample were measured between the true and predicted profiles using Jenson-Shannon Divergence (y axis, JSD). For comparison, random guess error (blue line) is shown to indicate a lower performance boundary. Spikes in error rates above 0.1 JSD are mostly related to time points in which the species average coverage drops below 10×, preventing reliable SNP profiling (Supplementary Fig. 7b).