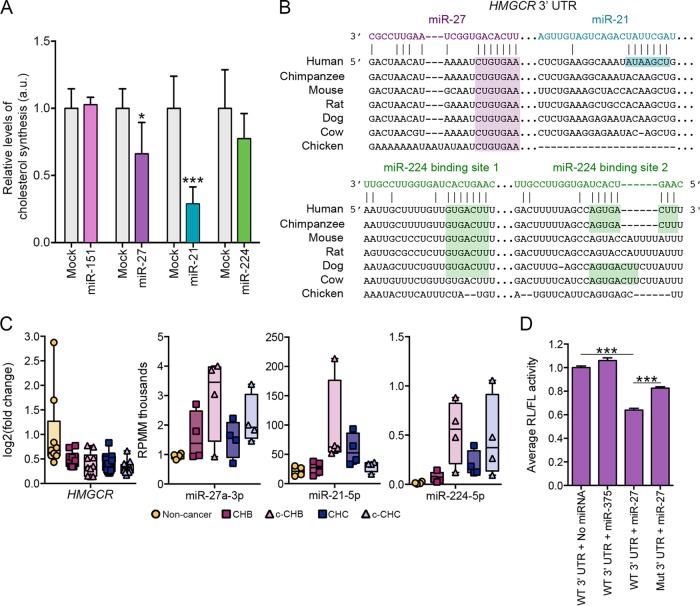
FIG 6 .
Candidate miRNA master regulators miR-27 and miR-21 control cholesterol synthesis in vitro. (A) Human hepatoma (Huh7) cells transfected with a 10 nM concentration of either miRNA mimic or transfection reagent alone (mock transfected) for the different RNAs: for miR-151, mock (n = 6) and mimic (n = 3); for miR-27, mock (n = 6) and mimic (n = 6); for miR-21, mock (n = 6) and mimic (n = 6); for miR-224, mock (n = 5) and mimic (n = 5). De novo cholesterol synthesis was measured by radiolabeled acetate incorporation assay (Materials and Methods). Relative change compared to mock transfection is shown. (B) Predicted target sites in the HMGCR 3′ UTR for miR-27, miR-21, and miR-224 as determined by the TargetScan algorithm. Gaps introduced to maximize alignment are indicated by dashes. (C) Relative levels of HMGCR mRNA, miR-21, miR-27, and miR-224 across the disease categories. Fold change values for HMGCR are based on comparison to the values for uninfected controls. RPMM, reads per million mapped reads. (D) Effects of miR-27 mimic (10 nM) and miR-375 (10 nM; negative control) on the activity of Renilla (RL) luciferase containing either wild-type (WT) or mutated (Mut) HMGCR 3′ UTR normalized to firefly luciferase (FL) in transfected Huh7 cells. Each condition was tested twice with three technical replicates each (total n = 6). The mutation was targeted to the predicted miR-27 target site. P values were determined using two-tailed unpaired Student’s t test and are indicated as follows: *, P < 0.05; ***, P < 0.005.