FIG 3 .
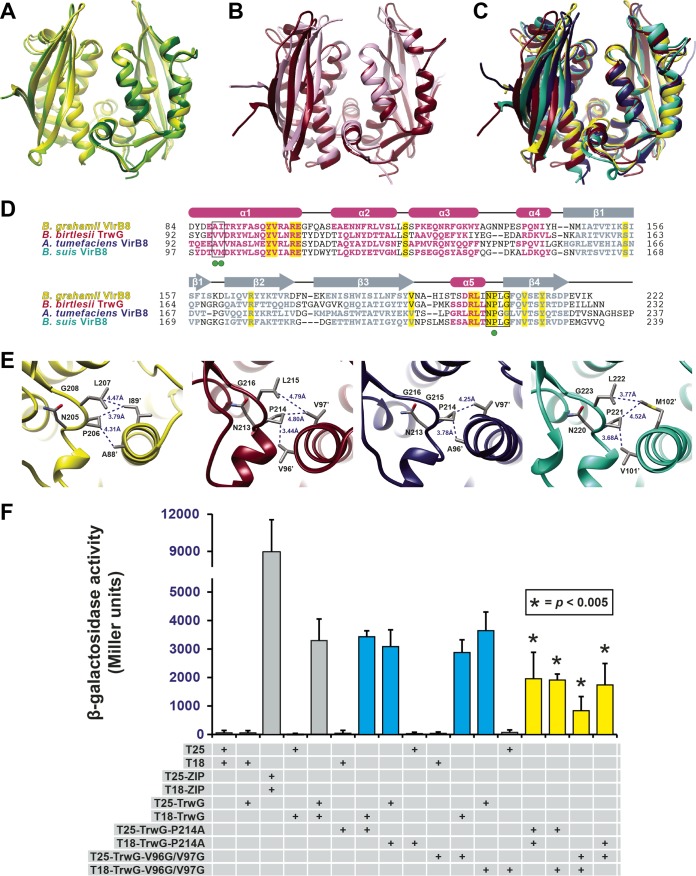
Bartonella VirB8 and TrwG proteins form structurally conserved homodimers. (A) Superimposition of the ribbon representations of the VirB8 crystal structures from B. quintana strain Toulouse (dark green) (PDB ID 4LSO), B. grahamii strain as4aup (yellow) (PDB ID 4KZ1) and B. tribocorum strain CIP 105476 (light green) (PDB ID 4MEI). (B) Superimposition of the ribbon representations of the TrwG crystal structures from B. birtlesii strain LL-WM9 (burgundy) (PDB ID 4JF8) and B. grahamii strain as4aup (pink) (PDB ID 4NHF). (C) Superimposition of the ribbon representations for the crystal structures of B. grahamii VirB8, B. birtlesii TrwG, Agrobacterium tumefaciens strain C58 VirB8 (dark blue) (PDB ID 2CC3) (44), and Brucella suis biovar 1 (strain 1330) VirB8 (aquamarine) (PDB ID 2BHM) (45). (D) Sequence alignment of VirB8/TrwG proteins and secondary structure assignment. Sequences were extracted from a larger alignment (see Text S4 in the supplemental material). Sequences are the globular domains depicted in panel C. For each protein, residues involved in the major dimerization site are boxed. Invariant residues are highlighted in yellow. Magenta bars and gray arrows depict the α-helices and β-strands for the VirB8/TrwG structure, with colored residues in the proteins corresponding to these structural features. Green dots mark the residues mutated within the dimerization interface of B. birtlesii TrwG. (E) High-resolution depiction of the major dimerization sites for the proteins illustrated in panels C and D. From left to right: B. grahamii VirB8, B. birtlesii TrwG, A. tumefaciens VirB8, and B. suis VirB8. (F) Analysis of B. birtlesii TrwG-TrwG interactions using the bacterial two-hybrid system. Different plasmid combinations were transformed into adenylate cyclase deficient and cAMP-specific-phosphodiesterase-deficient E. coli strain APE304. After overnight growth in liquid Luria-Bertani medium, β-galactosidase activity was measured and calculated (in Miller units). T25-ZIP and T18-ZIP are positive-interaction control plasmids encoding dimer-forming yeast transcription factor GCN4 (89). Values are from three independent experiments, all analyzed in triplicate. Blue bars depict wild-type-mutant interactions, while yellow bars depict mutant-mutant interactions.