FIG 6 .
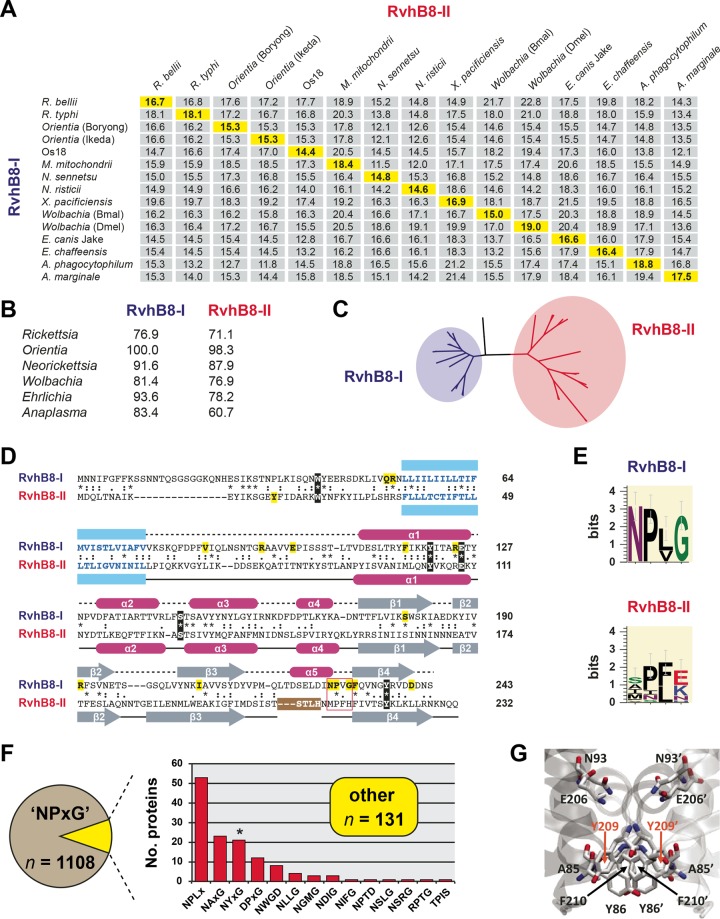
Conserved RvhB8-I and RvhB8-II paralogs are highly divergent from one another. (A) Pairwise divergence between RvhB8-I and RvhB8-II proteins from select rickettsial species. Numbers are amino acid identity (percent) as calculated across a global RvhB8 alignment (see the text for details). Highlighted values on the diagonal depict divergences between paralogs encoded within the same genome. Full species names and NCBI GenBank accession numbers for all proteins are provided in Text S1 in the supplemental material. (B) Across species and strains within the same genus, RvhB8-I is more conserved than RvhB8-II. Numbers are amino acid identity (percent) as described for panel A. Complete percent identity matrices used to estimate protein divergence are provided in Text S5 in the supplemental material. (C) Phylogeny estimation of RvhB8 proteins reveals higher divergence within the RvhB8-II clade than the RvhB8-I clade. ML-based phylogeny was estimated with RAxML on the unmasked global RvhB8 alignment (WAG + gamma + Ι). A complete tree, as well as phylogenies estimated from the masked alignment with other substitution models, is provided in Text S5 supplemental material. (D) Comparison of R. typhi RvhB8-I and RvhB8-II proteins. Sequences were aligned according to structure in SPDBV using “magic fit” followed by “improved fit” algorithms. The alignment shows predicted (top) and solved (bottom) structures for RvhB8-I and RvhB8-II, respectively. Predicted transmembrane-spanning regions (76) are colored blue. Five residues conserved across all RvhB8 proteins are in black, with residues conserved only in RvhB8-I (n = 15) or RvhB8-II (n = 1) highlighted in yellow (see Text S5 in the supplemental material). The NPXG motif is in a red box (see the description of panel E below). For RvhB8-II, the region of proteolysis (STLH) that occurred during crystallization is in a brown box. (E) Composition of the NPXG motif across 15 RvhB8-I (top) and 15 RvhB8-II (bottom) proteins. Sequence logos were generated using WebLogo v.3.3 (77). (F) Analysis of the conservation of the NPXG motif across 1,239 nonredundant proteobacterial VirB8 proteins (excluding Rickettsiales). Proteins lacking the conserved NPXG motif (10.6%) were placed in 14 categories based on their alternative sequences and ranked by their frequency (see Text S4 in the supplemental material for structural modeling of proteins within each category). (G) Example of a canonical interaction across the NPXG motif for Yersinia pestis biovar microtus strain 91001 (NP_995427), which contains the alternative sequence NYFG.