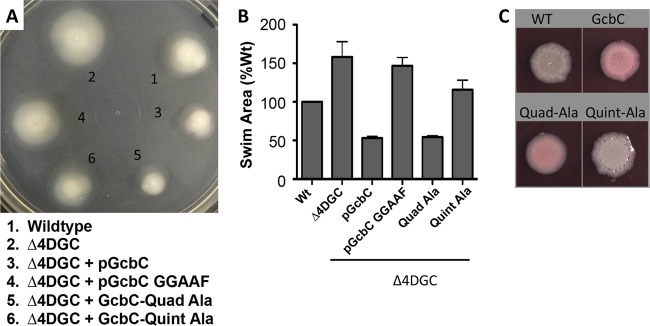
FIG 3 .
The α5GGDEF helix of GcbC contributes to promoting biofilm formation but is not required for GcbC swim motility. (A) Swim phenotypes of wild-type P. fluorescens, the Δ4DGC mutant, and the Δ4DGC mutant expressing GcbC, GcbC GGAAF, or the quadruple (Quad-Ala) and quintuple (Quint-Ala) alanine mutant variants. Strains were grown overnight in LB and normalized to cell density, and 100 µl was dispensed into a 96-well plate. A 200-µl pipette tip was dipped into each well and plunged into an 0.35% agar K10 minimal medium plate supplemented with 0.1% arabinose. Plates were incubated at 30°C for 36 h and photographed. (B) Quantified swim area from experiments performed as in panel A. Error bars show standard deviations from 3 biological replicates which were generated from 3 technical replicates per biological replicate. Swim area was normalized to the wild-type (Wt) strain. (C) The quadruple and quintuple alanine mutants of GcbC were analyzed by Congo red binding by transforming the plasmids into P. aeruginosa. The binding of the red pigment by these P. aeruginosa strains indicates exopolysaccharide production, which serves as a surrogate for the level of c-di-GMP produced. The quadruple mutant (Quad-Ala) shows similar levels of Congo red binding as the wild-type GcbC, while the quintuple mutant (Quint-Ala) shows reduced binding compared to the strain expressing wild-type GcbC. The wild-type (WT) P. aeruginosa PA14 strain served as a control, and strains were grown for 36 h.