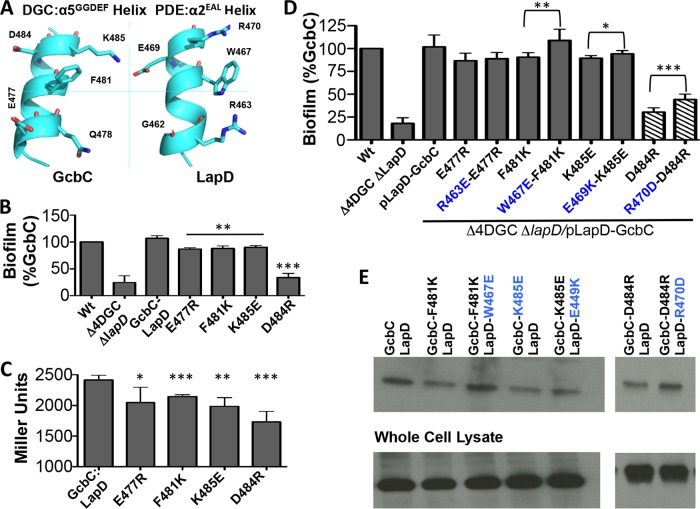
FIG 4 .
The α5GGDEF helix of GcbC interacts with a helix on LapD. (A) The regions of GcbC and LapD that may interact with each other are found on α-helices. Five surface residues that are possible points of contact between the GcbC helix and the LapD helix are shown in this ribbon diagram. (B) Nonconservative mutations in GcbC’s interaction surface were assayed for biofilm formation using the microtiter well assay at 6 h. **, P < 0.01; ***, P < 0.001 (compared to the control strain expressing wild-type GcbC). Error bars show standard deviations from 3 biological replicates which were generated from 8 technical replicates per biological replicate. Biofilm formation was normalized to the wild-type strain and is displayed as percent biofilm induction relative to the strain expressing wild-type GcbC. (C) The same mutations as those shown in panel B were assessed for interaction using a B2H experiment, confirming that GcbC’s α5GGDEF helix contributes to full interaction with LapD. *, P < 0.05; **, P < 0.005; ***, P < 0.001 (compared to interaction between wild-type GcbC and LapD). Error bars show standard deviations from 4 biological replicates which were generated from 3 technical replicates per biological replicate. (D) The disruptive GcbC mutations used in panel B were paired with potential compensatory mutations on LapD and assayed for biofilm formation. Suppression of biofilm disruption was observed for several pairs of mutants. To keep the final pair of mutants in the linear range of the assay, the D484R mutation was induced with 0.1% arabinose as indicated by the hatched bars. *, P < 0.05; **, P < 0.005; ***, P < 0.0005 (for each suppressor mutant relative to its unsuppressed GcbC mutant). Error bars show standard deviations from 7 biological replicates which were generated from 8 technical replicates per biological replicate. Biofilm formation was normalized to the wild-type strain and is displayed as percent biofilm induction relative to GcbC. (E) Compensatory LapD α2EAL helix mutations restore interaction with GcbC α5GGDEF helix mutations. Shown is a Western blot following a coprecipitation assay where cells expressing a 6-histidine-tagged LapD and HA-tagged GcbC were lysed, LapD-6H was immobilized on cobalt resin, and beads were washed before blotting with an anti-HA antibody. The bottom of the panel indicates the level of protein in the whole-cell lysate used for the pulldown assays.