Figure 2.
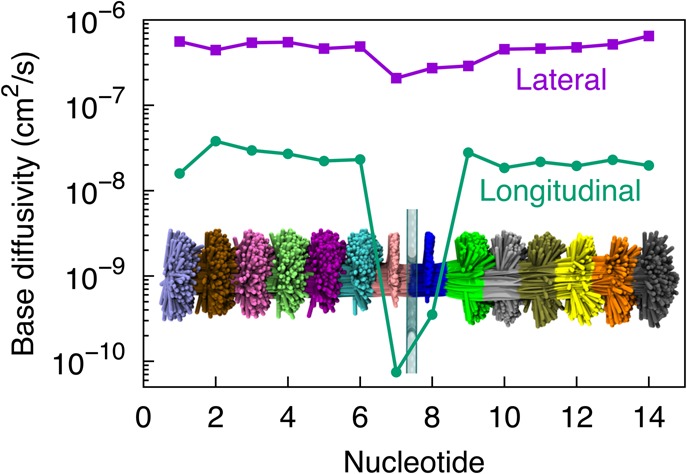
Fluctuations of ssDNA nucleotides. The lateral (parallel to graphene) and longitudinal (normal to graphene) diffusivities (defined in Methods) for nucleotides of poly(dA) ssDNA shown were evaluated from the last 100 ns of a 120 ns equilibration simulation. Nucleotides numbered 7 and 8 with lower longitudinal fluctuations are those that adhere to the graphene layer as shown in Figure 1b (bottom); nucleotides 1–6 and 9–14 that are not in direct contact with the graphene layer exhibit extensive fluctuations. The inset shows overlapped conformations of ssDNA in a 100 ns MD trajectory at 100 ps intervals; in comparison to Figure 1, where ssDNA is oriented along the vertical axis, in the inset here, ssDNA is shown oriented along the horizontal to better fit into the graph.
