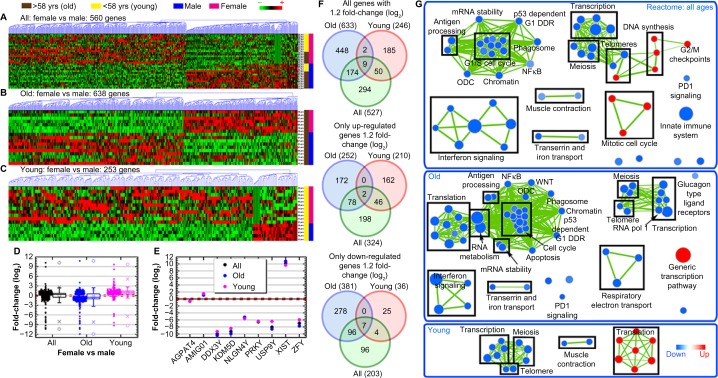
Figure 2.
Global gene regulation changes as a function of age and gender. Hierarchical clustering of genes by average linkage (UPGMA) and Euclidean distance calculation from the DLBCL TCGA mRNA data with a t-test, P-value < 0.05, for (A) female-to-male patient comparisons combining all ages, (B) old patient (>58 years old) only comparisons for female and male patients, and (C) young patient (<58 years old) comparison for female and male patients. (D) The average mRNA signal log2 fold-change comparing age-independent analysis to young and old-specific analyses. Whiskers show the range of the outliers, with max and min values as Ο and the 1 and 99th percentile outliers as X. (E) The average signal log2 fold-change for the nine common genes between the three different sets of analysis. (F) Venn diagrams of the genes with 1.2 fold-change for comparisons between age-independent analysis and old and young-specific analyses with separate Venn diagrams for only the upregulated and downregulated genes. The numbers in the parenthesis indicate the overall number of genes in each group. (G) Network representation of only Reactome-related gene sets from GSEA C2 gene set annotations. Leading edge analysis with an FDR < 0.05 determined significant gene sets enriched for each group. The size of each node reflects the amount of molecules involved in each gene set. The edge (green lines) represents the number of genes associated with the overlap of two gene sets (or nodes) that the edge connects. Clusters were named according to common function in each grouping. Upregulated gene sets are denoted by red color and downregulated gene sets are denoted by blue color.