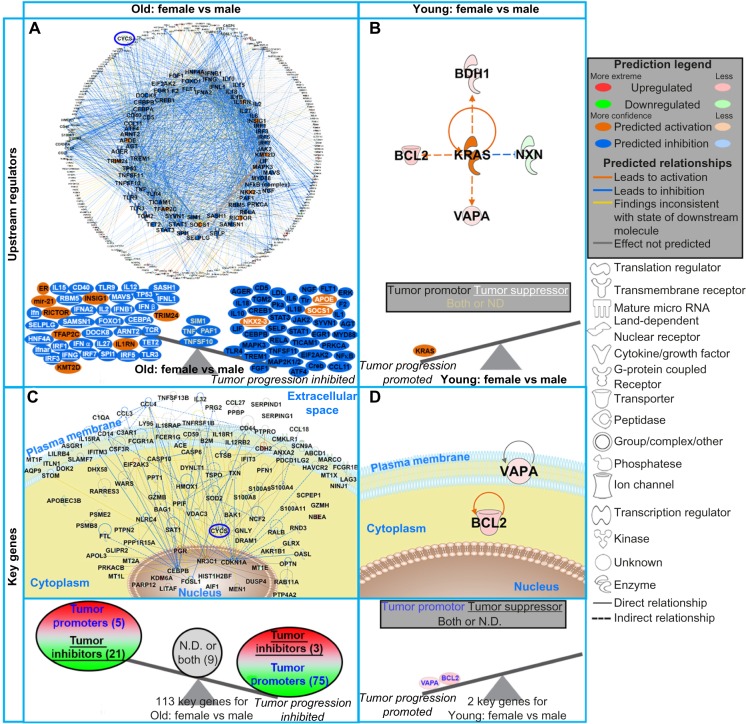
Figure 4.
The impact of predicted upstream regulators and key genes from DLBCL patients on tumor progression. (A and B) The top panel shows network representation of the upstream regulators for each type of analysis with the genes involved for each prediction. The second panel shows schematic of the activation states of the upstream regulators from Supplementary Table 2, illustrating the balance between the tumor promoters (black text) and tumor suppressors (white text and underlined) with a predicted activation (orange) or predicted inhibition (blue). The upstream regulators with both promoter and suppressor effects or ND effects are shown in the middle of the balance (yellow text). (C and D) The third panel shows gene network analysis for the key genes involved in each type of analysis. The pathway analysis was done with IPA software. The predicted relationships between all genes are also shown. Log2 fold-changes (with a cutoff of 1.2 fold-change) to the gene expression were used to obtain different shades of green for fold-change in downregulated genes, while different shades of red depict fold-changes in upregulated genes. The darker the shade of green or red, the greater the fold-change. The bottom panel shows a schematic of the key significant genes (Supplementary Table 4), determined to be significant in regulating many functions for each comparison, illustrating the balance between the tumor promoters (blue text) and tumor suppressors (black text and underlined) with the log2 fold-change color coded as before. Genes with both promoter and suppressor effects or ND effects are shown in the middle of the balance (black text). CYCS is circled in blue throughout the networks. In panel (C), the individual genes are not shown due to the quantity of key genes. For a complete list of key genes, see Supplementary Table 4.