Fig. 1.
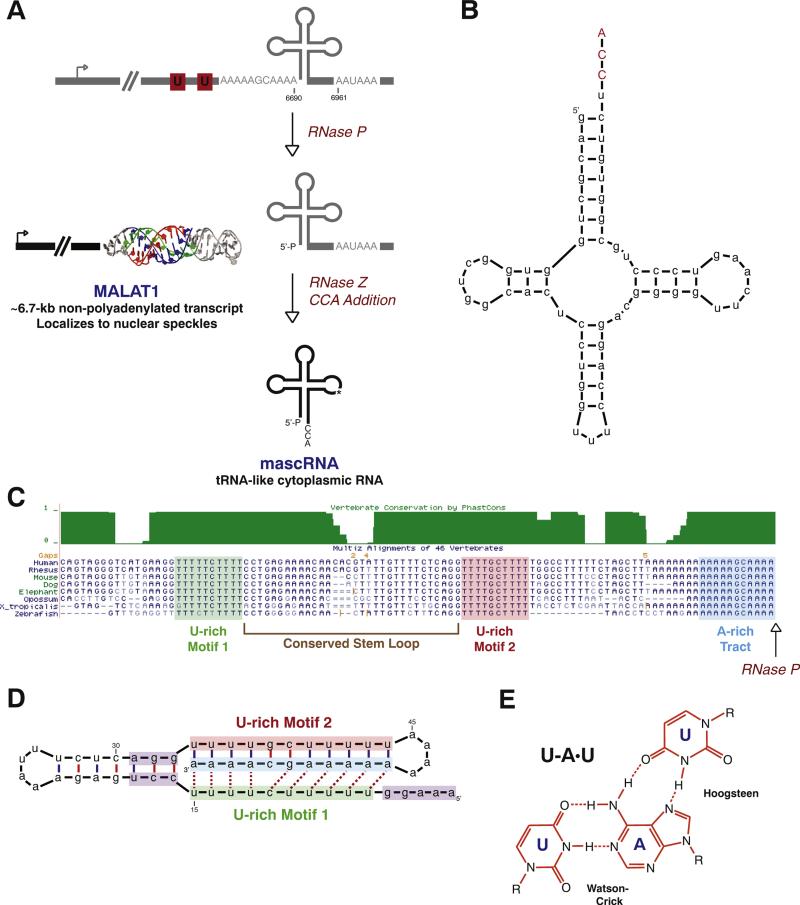
The MALAT1 locus generates a nuclear-retained long noncoding RNA and a tRNA-like small RNA. (A) Rather than using the canonical cleavage/polyadenylation machinery, the 3′ end of MALAT1 is almost always generated by tRNA biogenesis factors. First, RNase P cleavage simultaneously generates the mature 3′ end of MALAT1 and the 5′ end of mascRNA. The tRNA-like small RNA is subsequently cleaved by RNase Z, subjected to CCA addition, and exported to the cytoplasm. In contrast, the mature MALAT1 transcript localizes to nuclear speckles and its 3′ terminus is protected by a triple helical structure (PDB code 4PLX [84]). The mouse coordinates (GenBank accession number FJ209304) are given. (B) Mouse mascRNA adopts a tRNA-like cloverleaf secondary structure. CCA (denoted in red) is post-transcriptionally added by the CCA-adding enzyme. (C) As shown by the Multiz Alignment track of the UCSC Genome Browser, highly conserved A- and U-rich tracts are present immediately upstream of the MALAT1 RNase P cleavage site (denoted by arrow). (D) These conserved sequence motifs form base triplets (denoted by dashed lines) that protect the 3′ end of MALAT1 from degradation. A minimal triple helix that supports both RNA stability and translation [82] is shown. Nucleotides that function in promoting translation are denoted in purple. (E) U–A·U base triplets form via Hoogsteen hydrogen bonds to the major groove of a Watson–Crick base-paired helix.