Fig. 3.
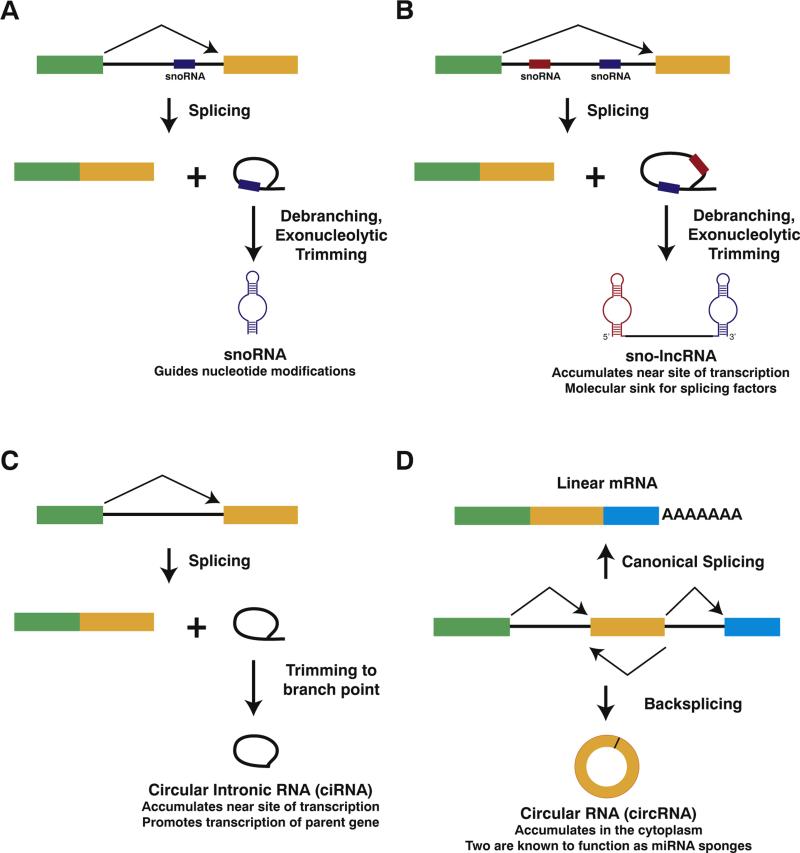
The pre-mRNA splicing machinery generates a number of non-polyadenylated noncoding RNAs. (A) When a snoRNA sequence is encoded in an intron, pre-mRNA splicing releases the excised intron, which is subsequently debranched and trimmed to produce the mature ~70–200 nt snoRNA. In this case, a box C/D snoRNA is shown. (B) In contrast, when two snoRNA sequences are present in a single intron, debranching and trimming of the excised intron produce a long noncoding RNA with snoRNA ends. (C) After splicing, some introns fail to be debranched and accumulate as stable circular intronic RNAs in the nucleus. These transcripts are covalent circles due to the 2′,5′-phosphodiester bond between the 5′ end of the intron and the branch point adenosine. (D) Pre-mRNA splicing can generate linear or circular RNAs comprised of exons. If the splice sites are joined in the canonical order, a mature linear mRNA is generated that is subsequently polyadenylated (top). Alternatively, the splicing machinery can backsplice and join a splice donor to an upstream splice acceptor, generating a circular RNA whose ends are covalently linked by a 3′,5′-phosphodiester bond (bottom).