Figure 2.
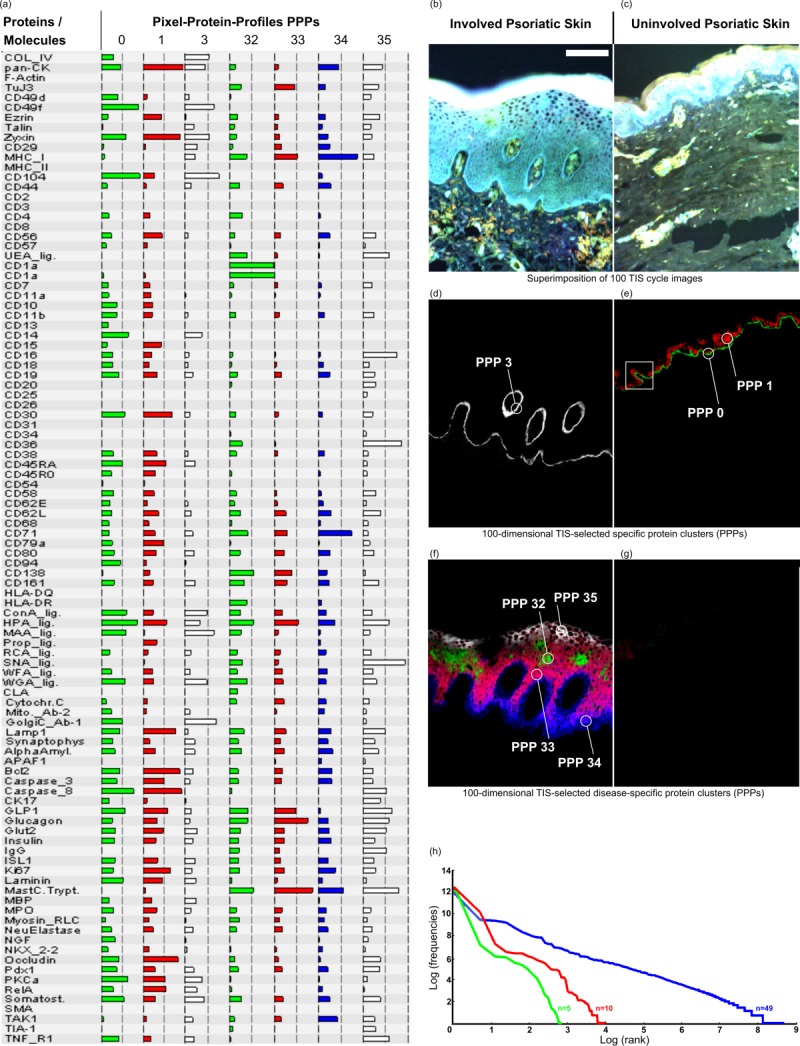
Discovery of disease specific 100-dimansional protein profiles simultaneously and in real time in morphology intact tissue sections at a PCMD of 256100 per pixel, exemplified in human skin. (a) List of 100 co-mapped biomolecules and selected protein profiles (0, 1, 3, 32 – 35) specific for diseased (d, f) and normal skin (e, g). (b,c) Diseased (b) and normal skin (c) are highlighted by pseudo colouring as histological stain for morphological orientation. Note that, by moving the cursor over the pixels, the software directly recognises in realtime which protein profiles are specific for the diseased (d, f) or normal skin (e, g). For example, pixel protein profiles (PPP) with numbers 0 and 1 are specific for the normal skin (e), and PPP 3, as well as PPPs 32 – 35 (d, f, respectively) are specific for the diseased skin. Note: For many similar applications at real time see webpage of the human toponome (HUTO) project (www.huto.toposnomos.com). (h) Power law (Zipf’s law) substantiates highly organised protein systems, as seen in (d-g). If 49 molecules are co-mapped Zipf’s law applies (blue line), but does not apply, if <15 molecules are co-mapped (red and green lines). This is revealed by plotting the log-log-relationship of thousands of distinct protein assemblies in toponome data sets 5,6. Bar: 100 µm (b–g). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
