Abstract
Background and Objective:
Haemophilus influenzae type b (Hib) is divided into two distinct genotypes, type I and type II, based on the structure of capsular polysaccharides. The capsulation locus of Haemophilus influenzae type b consists of three functionally distinct regions, designated regions 1 to 3. Region III contains hcsA and hcsB genes; however, notable sequence variation in this region can be used to recognize different Hib genotypes. The purpose of this study was to investigate the prevalence and genotype of the Hib strains isolated from patients with invasive disease in Iran.
Materials and Methods:
In the present study, 8 pairs of primers were used for identification and serotyping of encapsulated Haemophilus influenzae strains, as well as confirmation of species identification. Additionally, in order to identify the capsular genotypes of Haemophilus influenzae type b (type I and II), two additional primer pairs were used to amplify the hcsA gene.
Results:
Out of 50 isolates of H. influenzae, four were found to be type b. Interestingly, among these 4 Hib isolates, 2 strains belonged to the type-II category.
Conclusion:
Our study shows that the prevalence of both Hib types I and II seems to be high in Iran.
Keywords: Hib, genotype, hcsA, Iranian patients
INTRODUCTION
Haemophilus influenzae is a Gram-negative, coccobacillary, facultatively anaerobic bacterium belonging to the Pasteurellaceae family. Haemophilus influenzae strains, which commonly reside as commensals within the human pharynx and may cause respiratory or invasive infections, are variable for the presence of a polysaccharide capsule (1). Isolates of H. influenzae can be subdivided into two major forms, encapsulated and non-encapsulated. Encapsulated strains express one of six structurally and antigenically distinct capsular polysaccharides, referred to as serotypes a through f (2). While the capsules are generally composed of a polysaccharide containing two hexose sugars as subunit carbohydrates, the type b polysaccharide capsule is the only capsular type which contains two pentose monosaccharides rather than the hexose sugars as subunit carbohydrates (3, 4). More importantly, Haemophilus influenzae type b (Hib) conjugate vaccines which consist of the capsular polysaccharide of Hib, polyribosyl-ribitol-phosphate (PRP), conjugated to a carrier protein are safe and highly effective in preventing invasive Hib disease (5). In the pre-vaccination era, about 90% of the invasive cases, such as meningitis and sepsis, were associated with type b strains (6). Since the introduction of the Hib conjugate vaccine in Europe and America in the 1990s, the incidence of invasive Hib disease has dramatically decreased in most countries (5).
Based on genetic analyses, two major phylogenetic divisions of Encapsulated H. influenzae, divisions I and II, have been already described (7–9). Division I consists of the great majority of serotype a and b strains and all of serotype c, d, and e strains. On the other hand, division II includes all strains with serotype f and some with serotypes a and b (7, 9, 10). It should be noted that most isolates of H. influenzae from clinical infections belong to clonal division I (7, 11). All encapsulated H. influenzae strains, whether division I or II, contain common genes for the production of their respective polysaccharide capsules (cap genes) found within the cap locus that is divided into three functionally distinct regions (9, 10). Region I contains four genes, bexA to bexD, which play a key role in capsular polysaccharide export. Region II, flanked by regions I and III, includes the genes bcs1 to bcs4 involved in the biosynthesis of the serotype-specific polysaccharide in Hib (9, 12, 13). Region III comprises genes referred to as hcsA and hcsB, which share significant homology with genes in a number of other encapsulated pathogens, including Neisseria meningitidis (lipA and lipB), Escherichia coli K1 and K5 (kpsC and kpsS), Mannheimia (Pasteurella) haemolytica (wbrA and wbrB), P. multocida A:1 (phyA and phyB), P. multocida B:2 (wcbA and wcbO), Burkholderia mallei and Burkholderia pseudomallei. Collectively, the Haemophilus influenzae type b hcsA and hcsB gene products have complementary functions involved in the transport of capsular polysaccharide across the outer membrane and are essential for virulence. Surprisingly, the hcsA and hcsB gene products may be viable targets for antimicrobial development. In division I type b strains, these three loci are flanked by three 771 bp insertion elements, IS1016, whereas the capsulation locus in division II type b strains is not flanked by IS1016. The presence of the insertion element IS1016 increases the copy number of the cap locus. In fact, an increase in the copy number (up to 6 copies per cell) has been detected in clinical isolates (14, 15). The aim of this study was to determine the genotypes of Hib strains isolated from clinical samples, and to explore strategies to improve vaccine efficacy.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
A total of 50 clinical isolates of H. influenzae originated from blood, nasopharynx, eye, CSF and bronchoalveolar lavage fluid from hospitalized patients were used in the current study. All isolates were collected during the years 2010 to 2012 from Milad, Imam Khomeini and Mofid hospitals in Tehran, Iran. They were examined for morphological characters, biochemical reactions and the requirements for X and V factors (16). The samples were cultured on chocolate agar plates, and shipped to the Department of Bacteriology, Pasteur Institute of Iran, Tehran. Afterwards, the H. influenzae strains were cultured on chocolate agar containing 10% sterile sheep blood, and then incubated overnight at 37°C in a humidified 5% CO2 incubator. The serotypes of the strains were also determined by polymerase chain reaction (PCR) as described by Falla et al. (17).
Extraction of bacterial DNA.
After 24 hours of incubation on chocolate agar, an inoculating loop of bacteria was gently scraped from each of the chocolate agar plates and individually suspended in 150 μl of double-distilled water (DDW). The mixture was briefly mixed on a vortex mixer, placed in a boiling water bath for 7 min and then centrifuged at 12,000 × g for 5 min. A 50-μl aliquot of the supernatant was transferred to a sterile tube and stored at −20°C until PCR testing (17).
Control strains.
H. influenzae type a (ATCC 9006), H. influenzae type c (ATCC 9007) and nontypeable H. influenzae (NTHi) (ATCC 49766) were used as positive control for genotype II, positive control for genotype I and negative control, respectively in PCR assays. The control samples were obtained from the microbial collection of the Department of Bacteriology, Pasteur Institute of Iran, Tehran, Iran.
Identification of H. influenzae species and capsular typing of encapsulated H. influenzae by PCR.
To confirm species identification, PCR amplification was performed to detect the omp6 gene from genomic DNA (18). Additionally, the PCR amplification was further carried out using the primer pair specific to bexA genes (HI-1 and HI-2) to distinguish encapsulated H. influenzae strains from NTHi strains. For capsular typing of encapsulated H. influenzae, six primer sets specific for capsule types a through f were used for all strains, which recognize sequences in capsule-specific genes located within region 2 of the cap locus (17). Isolates containing both the bexA gene and one of the cap-specific genes are designated as the specific capsule type. In contrast, isolates lacking both bexA and any of the cap genes are considered as NTHi, and those containing a cap gene but not bexA are designated as capsule-deficient H. influenzae type b or Hib− strains (19) (All primers are listed in Table 1).
Table 1.
The primer sets used in this study
| Primer name | Primer sequence (5′ to 3′) | Target | Amplicon size (bp) | Reference |
|---|---|---|---|---|
| F1-(F) | AACTTTTGGCGGTTACTCTG | Omp6 | 351 | 18 |
| R1-(R) | CTAACACTGCACGACGGTTT | |||
| HI-1 (F) | CGTTTGTATGATGTTGATCCAGAC | bexA | 343 | 17 |
| HI-2 (R) | TGTCCATGTCTTCAAAATGATG | |||
| a1 | CTACTCATTGCAGCATTTGC | a | 250 | 17 |
| a2 | GAATATGACCTGATCTTCTG | |||
| b1 | GCGAAAGTGAACTCTTATCTCTC | b | 480 | 17 |
| b2 | GCTTACGCTTCTATCTCGGTGAA | |||
| c1 | TCTGTGTAGATGATGGTTCA | c | 250 | 17 |
| c2 | CAGAGGCAAGCTATTAGTGA | |||
| d1 | TGATGACCGATACAACCTGT | d | 150 | 17 |
| d2 | TCCACTCTTCAAACCATTCT | |||
| e1 | GGTAACGAATGTAGTGGTAG | e | 1350 | 17 |
| e2 | GCTTTACTGTATAAGTCTAG | |||
| f1 | GCTACTATCAAGTCCAAATC | f | 450 | 17 |
| f2 | CGCAATTATGGAAGAAAGCT | |||
| HiHcsA12667F-I | GTACTTGTCATTGACCAAACTTT | hcsA-I | 450 | 23 |
| HiHcsA13116R-I | GGTATATTGAAAGTATGCTGCAT | |||
| HiHcsA12668F-II | TGCTTGTCATCGATCAAA | hcsA-II | 817 | 23 |
| HiHcsA13484R-II | ACTAAAGAAAGGGGTGCAA |
Capsular genotyping of Hib strains.
To determine the capsular genotypes, or phylogenetic relationships, of the Hib strains, PCR amplification was performed with two sets of primers designed by Leo Schouls et al (18). In this regard, two oligonucleotide primer pairs HiHcsA12667F-I and HiHcsA13116R-I, and HiHcsA12668F-II and HiHcsA13484R-II were used to detect capsular genotypes I and II of H. influenzae type b, respectively. The primers HiHcsA12667F-I and HiHcsA13116R-I permitted the amplification of a 450-bp DNA fragment, whereas another set of primers, HiHcsA12668F-II and HiHcsA13484RII, allowed the amplification of an 817-bp fragment (Table 1).
To discriminate between the two capsular genotypes, type I and type II, two separate PCR amplifications were performed for each H. influenzae type b strain. The PCR reactions were carried out in a final volume of 25 μL containing 2.5 μl of 10× PCR buffer, 1.5 μl of 25 mM MgCl2, 0.2 μL of 5 U/μL Taq DNA polymerase (Genet Bio, Korea), 0.5 μl of each 10 μM primer, 0.5 μL of 10 mM dNTP (Genet Bio Company, Korea), 17.3 μl of double-distilled water and 2 μL of genomic DNA. PCR conditions were as follows: an initial denaturation at 95°C for 15 min, 30 cycles of denaturation at 95°C for 30s; annealing at 52°C for 1 min; elongation at 72°C for 1min, and a final extension step at 72°C for 7 min.
Gel electrophoresis of PCR products.
6 μL of PCR products mixed with 2 μl of loading buffer were loaded into individual wells of 1% agarose gel, and electrophoresis was performed at 100 V for 45 min. Further, the sizes of the amplicons were measured by comparison with 3μl of the 100-bp DNA ladder (Pars Tous Biotechnology Company, Iran). The resulting PCR products were stained with ethidium bromide for 20 min and visualized under UV illumination. To confirm whether the amplified bands corresponded to hcsA type I or hcsA type II fragments, one of the PCR products from each gene fragment were randomly sequenced (TAG Copenhagen, Copenhagen, Denmark).
RESULTS
Preparation of samples.
A total of 50 isolates of H. influenzae, obtained from nasopharynx (n=39), CSF (n=2), blood (n=3) eyes (n=5) and bronchoalveolar lavage fluid (n=1), were examined (Table 2). All H. influenzae isolates were initially confirmed by culture and biochemical tests. The strains were then further characterized by PCR amplification of the omp6 gene to confirm the H. influenzae species. Moreover, the primer specific for the bexA gene was used to identify encapsulated strains of H. influenzae. In PCR assays, 4 (8%) of the 50 strains were able to amplify the bexA gene (a fragment of 343 bp in length). Capsular types (a–f) were determined for each of the 4 strains using specific primer pairs (Table 1). Out of 4 isolates of H. influenzae, 4 were typed as b, successfully amplifying the bcs3 gene (a fragment of 480 bp in length). Four strains of H. influenzae type b examined had been isolated from blood (2 patients) and CSF (2 patients) specimens. In addition, two samples had been obtained from the infants at the ages of 3 months (blood) and 1 year (CSF), but the age-specific data were not available about the two other samples (Table 2).
Table 2.
Characteristics of isolates used in this study
| Nomber of strains | P6 (Haemophilus influenzae) | bexA (type or nontype) | Serotype (a–f) | Genotype (I or II) |
|---|---|---|---|---|
| Nasopharynx 39 | + | - | No | No |
| CSF 2 | + | 2 (+) | 2 (b) | 2 (I) |
| Blood 3 | + | 2 (+), 1(−) | 2(b), 1(nontype) | 2 (II) |
| Eye 5 | + | - | No | No |
| *BAL 1 | + | - | No | No |
| Total 50 | 50 + | 4(type), 46(nontype) | 4(b), 0(non b) | 2(I), 2(II) |
Bronchoalveolar Lavage
Capsular genotyping of Hib strains.
As mentioned previously, H. influenzae type b strains are known to belong to both phylogenetic groups, clonal divisions I and II. In the present study, to determine which clinically isolated Hib strains belong to which phylogenetic groups, specific primers to the hcsA type I and hcsA type II genes were used to identify genotype I and II Hib strains, respectively. However, it is found that among 4 H. influenzae strains examined two strains belong to type I (Fig. 1), and the two other strains belong to type II (Fig. 2).
Fig. 1.
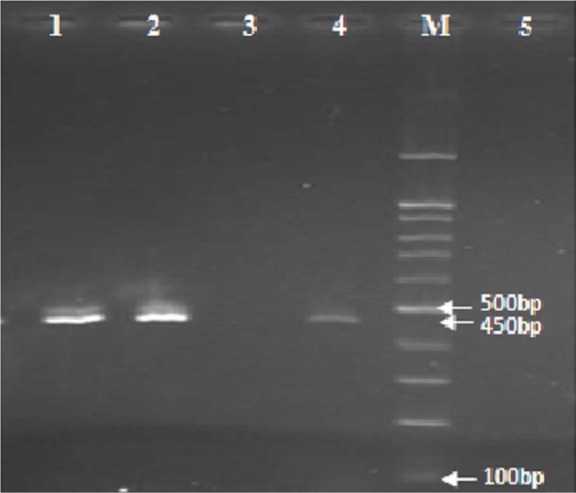
PCR amplification of the hcsA type I gene: the amplified DNA fragment was 450 bp in length, lane 1 and 2: clinical isolates of H. influenzae type b containing hcsA type I, Lane 3: clinical isolate of H. influenzae type b lacking hcsA type I, Lane 4: positive control containing H. influenzae type c (ATCC 9007), lane 5: negative control containing H. influenzae type a (ATCC 9006), M: Marker 100-bp.
Fig. 2.
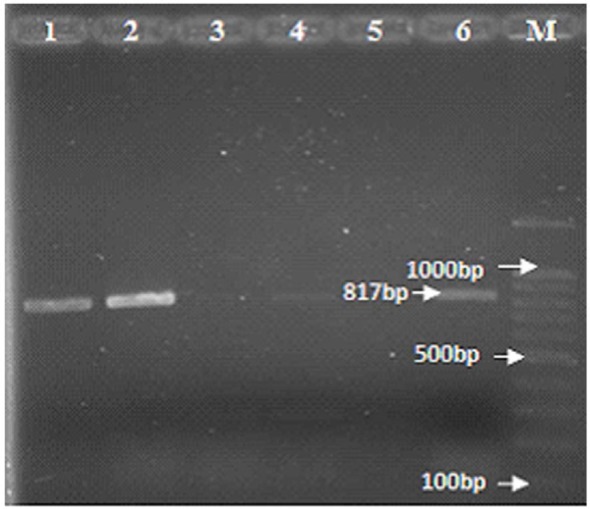
PCR amplification of the hcsA type II gene: the amplified DNA fragment was 817 bp in length, lane 1 and 2: clinical isolates of H. influenzae type b containing hcsA type II, Lane 3 and 4: clinical isolates of H. influenzae type b lacking hcsA type II, Lane 5: negative control containing H. influenzae type c (ATCC 9007), lane 6: positive control containing H. influenzae type a (ATCC 9006), M: Marker 100-bp.
Sequencing and BLAST homology search.
Two randomly selected PCR products of hcsA type I and hcsA type II genes were sequenced, confirming the identity of H. influenzae type b. After sequencing of PCR products, the newly determined sequences were analyzed using the Chromas software. In parallel, the nucleotide sequences of hcsA type I and hcsA type II were compared and aligned with those deposited in the GenBank database using the BLAST algorithm (http://blast.ncbi.nlm.nih.gov/). The nucleotide sequence data of hcsA type I and hcsA type II reported in this paper have been submitted to GenBank, and assigned the accession numbers KF263489 and KF263490 respectively.
DISCUSSION
Haemophilus influenzae is an environmentally sensitive and fastidious bacterium, making it difficult to isolate the pathogen from human clinical samples in microbiology laboratories. The diagnosis of bacterial meningitis is based on cerebrospinal fluid biochemical findings as well as clinical features. However, there is no specific treatment proven to be effective for bacterial meningitis, and patients are randomly assigned to the treatment regimens, often resulting in misdiagnosis and improper treatment. Hence, the isolation of even a single colony of H. influenzae from suspected cases will be valuable. H. influenzae type b remains one of the most common causes of bacterial meningitis in Iran as reported by the Iranian Ministry of Health. In Tehran (2005), among 32 CSF isolates of H. influenzae with positive cultrure, 5 strains(15.6%) were Hib (20). In another study in Tehran the rate of transmission of H. influenzae type b in children aged ≤5 was reported as 7.8% in 2007 (21) and recently in 2014, one study reported a 28% prevalence of H. influenzae among children ≤6 years old (22). Therefore, vaccination against H. influenzae type b will be included in the national immunisation program schedule to help control disease spread. In this light, the identification of H. influenzae serotypes is essential for vaccination strategies, supplied through domestic production or imports.
In this regard, two sets of primers were used to amplify the hcsA gene that could successfully discriminate between two capsular genotypes, type I and II. In 2008, Schools et al. revealed that the type I isolates carry more surface-bound capsular polysaccharides than type II isolates. The lower capsule production in type II strains may have resulted in a selective disadvantage, explaining the rapid disappearance of type II strains from the Dutch Hib population. Type II strains were isolated from 0–4 year old, non-vaccinated patients, suggesting that only type II strains are able to cause invasive disease in hosts who have not yet mounted an antibody response against the Hib capsule (23). Moreover, their results revealed that Hib strains isolated from patients in different geographic regions of the world show a considerably different distribution of the type I and II strains; In other words, the proportion of type II strains among the strains from the various global regions varied from 47% for strains from the African continent, 4% for Asian strains, 8% for Australia and 43% for Cuba. Furthermore, none of the Hib isolates from patients in other parts of Europe (Finland & Switzerland) belonged to the type II strains (23). On the other hands, the highest proportion of type II strains (73%) was found among Hib strains isolated from patients in the United States (23).
In the present study, 4 Hib strains isolated from Iranian patients were examined. The phenotypic and genotypic characteristics of Hib strains suggested that 2 Hib strains belonged to type I, and 2 other Hib strains were type II. It is important to note that 50% of Hib strains examined in this study belonged to the type-II category. However, this result is inconsistent with the results reported by other investigators carried out in many parts of the world, especially in Asia, because type II and type I Hib strains are completely dominant in the United States and in other countries in which the study was conducted, respectively. More importantly, these results demonstrate that the distribution pattern of Hib strains from Iran is closely similar to those from African countries and Cuba. To develop a broadly protective Hib vaccine, it would be better to use the native strains of type I PRP, because a recent study by Schouls et al. strongly demonstrated that type I strains produce PRP twice as much as type II stains. Notably, the purified PRPs should be used in the development of the Hib vaccine; this leads to reduce the time and cost for vaccine development and increase the extraction yield of PRPs. Sukupolvi-Petty et al. demonstrated that the products of the hcsA and hcsB genes facilitate transport of capsular polysaccharides across the outer membrane. Inactivation of hcsA alone resulted in accumulation of polysaccharides in the periplasm and a partial decrease in surface-associated polysaccharides, whereas inactivation of hcsB alone, or both hcsA and hcsB resulted in accumulation of polysaccharides in the periplasm as well as complete loss of surface associated polysaccharides. It is therefore feasible that alterations in the hcsA and hcsB genes would influence the degree of capsular polysaccharide export to the surface of the Hib cells, or the efficiency of retaining the exported polysaccharides on the surface (12). Meanwhile, after analyzing the complete sequences of the type I and type II Cap b loci, they showed that the variation between the capsular gene cluster sequences of type I and type II strains was predominantly found in the hcsA and hcsB genes. The nucleotide sequences of bex, bcs1 and bcs2 of type I and type II were identical (23). However, in another study, Kroll and Moxon compared capsule production in strains containing one or two copies of the cap b locus and observed a gene dosage effect, with twice as much polysaccharide capsule associated with two copies of the cap b locus. Interestingly, despite differences in capsule production, all strains were able to produce sustained bacteremia and meningitis in the infant rat model system, suggesting that only one copy of the cap b locus is sufficient for virulence in vivo. These findings suggest that a certain threshold level of polysaccharide capsule on the bacterial surface is required for intravascular survival and virulence of H. influenzae. However, despite the presence of less polysaccharide capsule on the surface of type II Hib strains, it seems that these strains can cause long-term meningitis in humans.
CONCLUSIONS
In summary, there are two different types of Haemophilus influenzae type b strains, which differ in coding sequences of genes involved in the synthesis of a polysaccharide capsule, especially the hcsA gene. This gene plays an important role in the transport of capsular polysaccharide to the cell surface. Importantly, any change or mutation in the sequence of this gene can affect on the amount of capsular polysaccharide present on the Hib surface as well as pathogenicity of H. influenzae in humans. In this study, it is found that the incidence and prevalence rates of both Hib types are significantly high in Iran. However, type I and type II Hib strains showed significant differences in their genotype (sequence diversity in the capsule-producing genes) and phenotype (the amount of capsular antigens) as well as pathogenicity. It seems that if type I Hib strains are used to produce the Hib vaccine, this vaccine can not only cover type II Hib strains but also induce a strong protection against H. influenzae type b infection, offering potential advantages such as economic benefits and high-quality products.
Acknowledgments
The present study was part of the M.Sc. dissertation of Somayeh Bagherzadeh khodashahri. This work was financially supported by the Grant No. 699 from Pasteur Institute of Iran, Tehran, Iran. We thank all members of the Bacteriology Department of Pasteur Institute of Iran.
REFERENCES
- 1. Davis GS, Sandstedt SA, Patel M, Marrs CF, Gilsdorf JR. Use of bexB to detect the capsule locus in Haemophilus influenzae. J Clin Microbiol 2011; 49: 2594–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pittman M. Variation and type specificity in the bacterial species Hemophilus Influenzae. J Exp Med 1931; 53: 471–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Waksman G, Caparon M, Hultgren S. (2005). Structural biology of bacterial pathogenesis. ASM Press; Washington DC. [Google Scholar]
- 4. Winn W, Allen S, Janda W, Koneman E, Procop G, Schreckenbeger P, et al. (2006). Koneman's Color Atlas and Textbook of Diagnostic Microbiology. 6nd ed Lippincott Williams and Wilkins, Philadelphia. [Google Scholar]
- 5. Ueno K, Nishi J, Imuta N, Tokuda K, Kawano Y. Presence of multiple copies of capsulation loci in invasive Haemophilus influenzae type b (Hib) strains in Japan before introduction of the Hib conjugate vaccine. Microbiol Immunol 2010; 54: 160– 163. [DOI] [PubMed] [Google Scholar]
- 6. Moloney AC, Fogarty I, Clarke P, Musser JM. Invasive Haemophilus influenzae in the Republic of Ireland. Eur J Clin Microbiol Infect Dis 1997; 16: 377– 380. [DOI] [PubMed] [Google Scholar]
- 7. Musser JM, Kroll JS, Granoff DM, Moxon ER, Brodeur BR, Campos J, et al. , Global genetic structure and molecular epidemiology of encapsulated Haemophilus influenzae. Rev Infect Dis 1990; 12: 75–111. [DOI] [PubMed] [Google Scholar]
- 8. Musser J, Kroll JS, Moxon ER, Selander RK. Evolutionary genetics of the encapsulated strains of Haemophilus influenzae. Proc Natl Acad Sci USA 1988; 85: 7758–7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Satola SW, Schirmer PL, Farley MM. Complete sequence of the Cap locus of Haemophilus influenzae serotype b and nonencapsulated b capsule-negative variants. Infect Immun 2003; 71: 3639– 3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kroll JS, Zamze S, Loynds BM, Moxon ER. Common organization of chromosomal loci for production of different capsular polysaccharides in Haemophilus influenzae. J Bacteriol 1989; 171: 3343–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhou J, Law D, Sill M, Tsang R. Nucleotide sequence diversity of the bexA gene in serotypeable Haemophilus influenzae strains recovered from invasive disease patients in Canada. J Clin Microbiol 2007; 45: 1996– 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sukupolvi-Petty S, Grass S, St Geme JW. The Haemophilus influenzae Type b hcsA and hcsB gene products facilitate transport of capsular polysaccharide across the outer membrane and are essential for virulence. J Bacteriol 2006; 188: 3870– 3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Van Eldere J, Brophy L, Loynds B, Celis P, Hancock I, Carman S, et al. Region II of the Haemophilus influenzae type be capsulation locus is involved in serotype-specific polysaccharide synthesis. Mol Microbiol 1995; 15: 107–118. [DOI] [PubMed] [Google Scholar]
- 14. Kroll JS, Loynds BM, Moxon ER. The Haemophilus influenzae capsulation gene cluster: a compound transposon. Mol Microbiol 1991; 5: 1549– 1560. [DOI] [PubMed] [Google Scholar]
- 15. Karlsson E, Melhus A. Nontypeable Haemophilus influenzae strains with the capsule-associated insertion element IS1016 may mimic encapsulated strains. APMIS; 2006. 114: 633–640. [DOI] [PubMed] [Google Scholar]
- 16. Kilian M. A taxonomic study of the genus Haemophilus, with the proposal of a new species. J Gen Microbiol 1976; 93: 9– 62. [DOI] [PubMed] [Google Scholar]
- 17. Falla TJ, Crook DW, Brophy LN, Maskell D, Kroll JS, Moxon ER. PCR for capsular typing of Haemophilus influenzae. J Clin Microbiol 1994; 32: 2382–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ueyama T, Kurono Y, Shirabe K, Takeshita M, Mogi G. High incidence of Haemophilus influenzae in nasopharyngeal secretions and middle ear effusions as detected by PCR. J Clin Microbiol 1995; 33: 1835–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mojgani N, Rahbar M, Taqizadeh M, Ashtiani MP, Mohammadzadeh M. Biotyping, capsular typing, and antibiotic resistance pattern of Haemophilus influenzae strains in Iran. Jpn J Infect Dis 2011; 64: 66– 68. [PubMed] [Google Scholar]
- 20. Taheri Kalani M, Akbari Nakhjavani F, Kazemi B, Bonakdar Hashemi F, Haghi Ashteiani M, Nouri K, et al. Survey rate isolation of Heamophilus influenzae type b in csf of pediatrics suspected to meningitis by culture and pcr in tehran center of medical children. TUMJ 2005; 63: 1006–1014(Persian). [Google Scholar]
- 21. Shiva F. Vaccination in children. SBMU 2007; 32: 1–4(Persian). [Google Scholar]
- 22. Jalali P, Mousavi SF, Rezaei N. Carriage rate of nasopharyngeal Haemophilus Influenzae among children under 6 Years Old in Tehran, Iran. J Med Microbiol Infect Dis 2014; 2: 23– 27. [Google Scholar]
- 23. Schouls L, van der Heide H, Witteveen S, Zomer B, van der Ende A, Burger M, et al. Two variants among Haemophilus influenzae serotype b strains with distinct bcs4, hcsA and hcsB genes display differences in expression of the polysaccharide capsule. BMC Microbiol 2008; 8: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]


