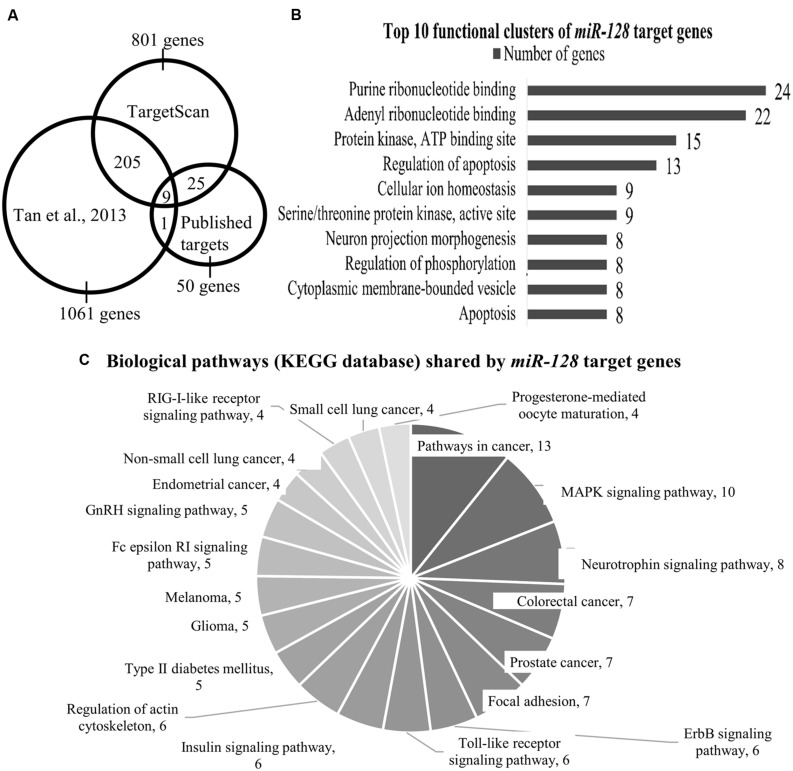
FIGURE 1.
(A) The Venn diagram shows the number of miR-128 target genes as identified by Tan et al. (2013) TargetScan and published targets. Total number of genes that are shared between Tan et al. (2013) and TargetScan is 214 genes; 34 genes are shared by TargetScan and our list of published miR-128 targets. (B) Top 10 functional clusters of miR-128 target genes as classified by DAVID bioinformatics tool. Using the mouse genome as the background, the threshold was set at the following criteria: classification stringency = highest; maximum EASE score = 0.05. EASE score is a modified Fisher Exact p value which range from 0 to 1, with 0 shows perfect enrichment. (C) Biological pathways (KEGG database) shared by miR-128 target genes as annotated by the KEGG pathway database. Using the mouse genome as background, the threshold was set at the following criteria: minimum number of genes for the corresponding pathway = 2; maximum EASE score = 0.05.