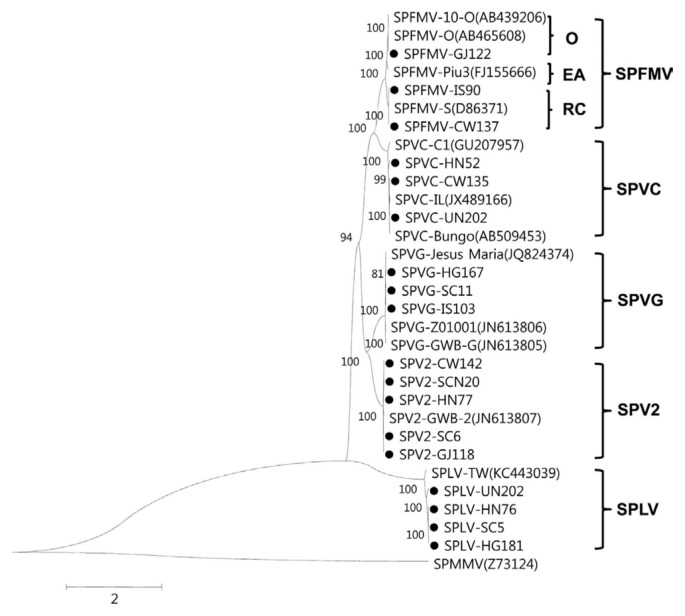
Fig. 5.
Phylogenetic trees reconstructed using the complete nucleotide sequences of the sweet potato potyvirus isolates. Phylogenetic trees were reconstructed using the maximum-likelihood method in MEGA 6. Numbers at each node indicate bootstrap percentages based on 1,000 replicates in maximum-likelihood tree. Horizontal branch length is drawn to scale with the bar indicating 2 nt replacements per site. SPMMV(Z73124) was used as the outgroup. Sweet potato feathery mottle virus (SPFMV), Sweet potato virus C (SPVC), Sweet potato virus G (SPVG), Sweet potato virus 2 (SPV2), Sweet potato latent virus (SPLV).