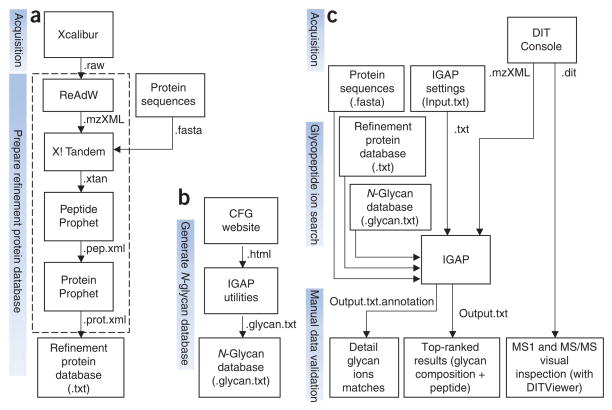
Figure 6.
Data analysis overview. (a) The refinement protein database is created from the protein IDs confidently identified with the LC-ESI-MS/MS data. Each ID is recorded on a separate line. This database is experiment specific. Protein identification may be performed using other search engines. (b) The N-glycan database is constructed with the data from Consortium for Functional Glycomics (CFG) Glycan Structures Database. This database only needs to be updated when new entries are found at CFG. (c) MALDI-DIT MS/MS spectrum is searched by IGAP, which generates the top 20 glycan composition results in the tab-delimited file ‘Output.txt’ and the matched peaks annotation in ‘Output.txt.annotation’. Manual validation of the results is performed. DITViewer provides visual access to the acquired spectra.