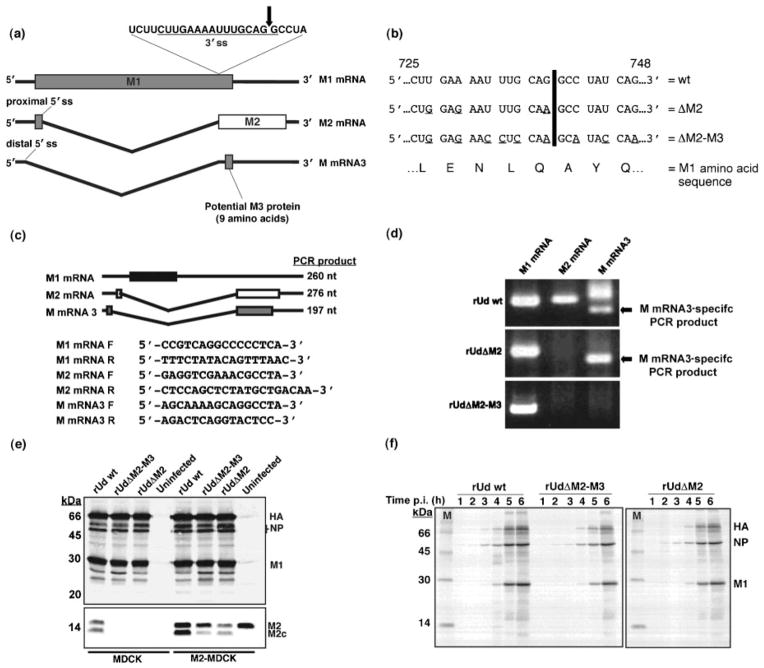
Fig. 1.
Generation and characterization of mutant viruses. (a) The splicing events of influenza A virus RNA segment 7. The arrow indicates the point of cleavage in the 3′ splice site (ss). (b) Nucleotide point mutations (underlined) introduced into the 3′ splice site of RNA segment 7, encoded on the pHH-M ‘rescue plasmid’, by site-directed mutagenesis. Numbers represent nucleotide position. The black line indicates the point of cleavage as marked in (a). (c) Schematic representation of segment 7-specific RT-PCR products. Primers were designed to allow the amplification of M1 mRNA- (black box), M2 mRNA- (white box) and M mRNA3- (grey box) specific PCR products (260, 276 and 197 nt, respectively). The primers for amplifying M2 mRNA and M mRNA3 products were designed to span the splice site junction and were based on those previously described by Cheung et al. (2005). Forward and reverse primers are indicated by F and R, respectively. (d) RT-PCR analysis of RNA segment 7-specific mRNAs in wt (rUd wt) and mutant (rUdΔM2 and rUdΔM2-M3) virus-infected MDCK cells. In the M mRNA3-specific lanes, the faster-migrating band is the M mRNA3-specific product and the slower migrating band is a product derived from M2 mRNA due to primer sequence similarities. (e, f) Analysis of viral protein synthesis in wt- and mutant virus-infected MDCK or M2-MDCK cells by Western blot (e) and radioimmunoprecipitation (1–6 h p.i.) (f) using an anti-Udorn antibody. Images were captured using Image Gauge version 3.3 software (Fuji Medical Systems).