Secretory and alga-specific genes are induced during gamete and zygote development in Chlamydomonas reinhardtii, concurrent with a dramatic increase in chloroplast cytosine methylation.
Abstract
The green alga Chlamydomonas reinhardtii undergoes gametogenesis and mating upon nitrogen starvation. While the steps involved in its sexual reproductive cycle have been extensively characterized, the genome-wide transcriptional and epigenetic changes underlying different life cycle stages have yet to be fully described. Here, we performed transcriptome and methylome sequencing to quantify expression and DNA methylation from vegetative and gametic cells of each mating type and from zygotes. We identified 361 gametic genes with mating type-specific expression patterns and 627 genes that are specifically induced in zygotes; furthermore, these sex-related gene sets were enriched for secretory pathway and alga-specific genes. We also examined the C. reinhardtii nuclear methylation map with base-level resolution at different life cycle stages. Despite having low global levels of nuclear methylation, we detected 23 hypermethylated loci in gene-poor, repeat-rich regions. We observed mating type-specific differences in chloroplast DNA methylation levels in plus versus minus mating type gametes followed by chloroplast DNA hypermethylation in zygotes. Lastly, we examined the expression of candidate DNA methyltransferases and found three, DMT1a, DMT1b, and DMT4, that are differentially expressed during the life cycle and are candidate DNA methylases. The expression and methylation data we present provide insight into cell type-specific transcriptional and epigenetic programs during key stages of the C. reinhardtii life cycle.
Chlamydomonas reinhardtii is a unicellular, biflagellate species of green alga found primarily in fresh water and soil (Harris et al., 2009). C. reinhardtii is an important reference organism for diverse eukaryotic cellular and metabolic processes, including photosynthetic biology (Rochaix, 2001), flagellar function and biogenesis (Silflow and Lefebvre, 2001), nutrient homeostasis (Grossman, 2000; Merchant et al., 2006; Glaesener et al., 2013), and sexual cycles (Goodenough et al., 2007). The nuclear and chloroplast genomes of C. reinhardtii have been fully sequenced, enabling genomic and epigenomic analyses (Maul et al., 2002; Merchant et al., 2007). The approximately 112-Mb haploid C. reinhardtii nuclear genome comprises 17 chromosomes. The circular chloroplast DNA (cpDNA) genome is 203 kb and present in 80 to 100 copies per cell that are organized into eight to 10 nucleoprotein complexes called nucleoids, which are distributed through the stroma.
Like many unicellular eukaryotes, C. reinhardtii has a biphasic life cycle where haploid cells can reproduce vegetatively by mitotic division or, alternatively, undergo a sexual cycle. Vegetative cells can propagate indefinitely when provided with nutrients and light. Upon nitrogen starvation, however, cells stop dividing and differentiate into gametes whose mating type (plus or minus) is determined genetically by an approximately 300-kb mating type locus on chromosome 6, with two haplotypes, MT+ and MT− (Umen, 2011; De Hoff et al., 2013). Gametes express a set of mating-related proteins that are different between minus and plus cells and that allow cells of opposite mating type to recognize each other and fuse to form a quadriflagellate zygote. Upon fertilization, the heterodimeric KNOX/BELL-type homeodomain proteins gamete-specific minus (GSM1) and gamete-specific plus (GSP1) initiate a zygote-specific developmental program that includes flagellar resorption, fusion of organelles including nuclei and chloroplasts, destruction of MT− cpDNA, and secretion of a thick, environment-resistant cell wall that protects the zygospore from cold, desiccation, and other environmental stresses (Cavalier-Smith, 1976; Catt, 1979; Grief et al., 1987; Brawley and Johnson, 1992; Goodenough et al., 2007; Lee et al., 2008). Upon return to favorable conditions of light and nutrients, zygospores undergo meiosis to produce four haploid progeny (two MT+ and two MT−) that can reenter the vegetative life cycle. While nuclear loci segregate in a Mendelian pattern of 2:2, both chloroplast and mitochondrial genomes are inherited uniparentally, with cpDNA inherited from the MT+ parent and mitochondrial DNA from the MT− parent (Nakamura, 2010; Nishimura, 2010).
While previous high-throughput expression studies have focused on the transcriptional programs underlying processes such as nutrient deprivation (Nguyen et al., 2008; González-Ballester et al., 2010; Toepel et al., 2011, 2013; Schmollinger et al., 2014), environmental responses (Simon et al., 2008, 2013; Matsuo et al., 2011; Fang et al., 2012), flagellar biogenesis (Albee et al., 2013), lipid accumulation (Miller et al., 2010; Boyle et al., 2012; Lv et al., 2013), and diurnal rhythms (Idoine et al., 2014; Panchy et al., 2014), only a few studies have explored the genome-wide transcriptional and epigenetic changes associated with the sexual cycle (Kubo et al., 2008; Ning et al., 2013; Aoyama et al., 2014). Several genes expressed in the early zygote, termed EZY genes, have predicted functions related to cell wall production, vesicular transport, and secretion (Ferris and Goodenough, 1987; Ferris et al., 2002; Kubo et al., 2008). A separate analysis of zygospore transcripts following light-induced germination revealed the up-regulation of photosynthetic and Met synthesis pathways (Aoyama et al., 2014).
DNA methylation studies have also been conducted on both the nuclear and chloroplast genomes (Hattman et al., 1978; Dyer, 1982). cpDNA methylation has been studied more extensively and shows dramatic changes in 5-methylcytosine (5meC) content at different stages of the C. reinhardtii life cycle. Vegetative cells have low levels of 5meC in cpDNA, while gametes show a substantial increase within cpDNA (12% 5meC in MT+ gamete cells and 4% in MT–; Royer and Sager, 1979; Feng and Chiang, 1984). In zygotes, MT− cpDNA is eliminated while MT+ cpDNA becomes hypermethylated. While differential cpDNA methylation was once thought to be part of a restriction-methylation system regulating uniparental inheritance (Burton et al., 1979), this model is unlikely, since loss of cpDNA methylation in MT+ cells does not result in its destruction in zygotes (Umen and Goodenough, 2001), and ectopic methylation of MT− cpDNA does not spare it from destruction (Bolen et al., 1982). However, previous studies are consistent with a role for 5meC in promoting cpDNA replication upon zygote germination, which can influence the amount of residual MT− cpDNA that is inherited by exceptional progeny (Umen and Goodenough, 2001; Nishiyama et al., 2004). An alternative proposed mechanism involves the digestion of MT− cpDNA by differentially localized or activated nucleases that are methylation insensitive early in zygote development before chloroplast fusion (Nishimura et al., 2002).
Several methyltransferase enzymes that modify cpDNA have been investigated biochemically (Sano et al., 1981), and one candidate chloroplast methyltransfersase gene has been cloned (Nishiyama et al., 2002, 2004). Since that time, the genome sequence of C. reinhardtii has become available (Merchant et al., 2007) and extensively annotated (Blaby et al., 2014) so that a comprehensive identification of genes encoding DNA methyltransferases can be undertaken.
Few studies have focused on the role of nuclear cytosine methylation in C. reinhardtii, but previous work has shown that induced silencing of nuclear transgenes does not correlate with transgene cytosine methylation levels, leaving open the question of what role cytosine methylation plays in chromatin structure and gene expression in C. reinhardtii (Cerutti et al., 1997). To date, the absence of detailed methylation maps has precluded a clear view of methylation patterns in the nuclear and chloroplast genomes of C. reinhardtii.
Here, we have performed RNA sequencing (RNA-seq)-based transcriptome analysis and bisulfite DNA sequencing of C. reinhardtii at different life cycle stages. We identify sex- and mating-related changes in gene expression, including genes that are preferentially expressed in gametes of each mating type or in zygotes. We generated a high-resolution map of nuclear and chloroplast cytosine methylation during the life cycle and identified candidate DNA methyltransferases whose expression profiles correlate with dynamic changes in cpDNA methylation patterns.
RESULTS
To quantify nuclear gene expression and 5-cytosine DNA methylation throughout the C. reinhardtii life cycle, we collected RNA and DNA samples at various stages for RNA and whole-genome bisulfite sequencing (Fig. 1). Samples were collected from vegetative and gametic cells of both mating types and zygotic stages to enable multiple comparisons. The design of the experiment was for matched DNA and RNA samples, but the RNA protocol was modified to avoid degradation. We note that for transcriptome studies, our vegetative samples were prepared in a different manner from those used for typical nitrogen-starvation studies. We grew all cultures to saturation on solid agar medium and then resuspended cells at high density (approximately 2 × 107 mL−1) under illumination in liquid medium (high-salt medium) with or without nitrogen for approximately 3 to 5 h. Under these conditions, neither culture grew measurably, but the cultures without nitrogen expressed the gametic program and efficiently mated at greater than 90% efficiency, while the cultures with nitrogen could not mate and did not express gametic marker genes (see below). The advantage of this procedure is that it minimizes the differences between the plus and minus nitrogen cultures that would normally be attributable to growth rate differences and thereby allows more reliable identification of mating-related gene expression. For DNA methylation studies, the vegetative and gametic samples were obtained from growing nitrogen-replete and nitrogen-starved samples, respectively, as described in “Materials and Methods.” Actively growing cultures were used for studies of methylation in vegetative cells, since agar plate-grown cells are already partially gametic and would require additional rounds of division in order to remove preexisting methylation.
Figure 1.
C. reinhardtii sexual life cycle and sequenced samples. Vegetative C. reinhardtii cells of each mating type (MT+ and MT−) can be induced to undergo gametogenesis by nitrogen starvation. Gametes of opposite mating type recognize each other through flagellar adhesion and fuse to form a diploid zygote. During zygote maturation, MT− cpDNA is eliminated, flagella are resorbed, and a thick zygote cell wall forms. Upon return to nitrogen and light, zygospores undergo meiosis to form four haploid progeny (two of each mating type, all containing uniparentally inherited MT+ parental cpDNA) that reenter the vegetative cycle. Colored boxes designate samples and material sequenced. Blue and red unfilled circles represent MT+ and MT– chloroplast genomes, respectively. Filled pink and light blue circles represent nuclear DNA from MT+ and MT– strains, respectively. BS-Seq, Bisulfite sequencing.
Sequence Polymorphisms between R3 (MT+) and CJU10 (MT–) Parental Strains
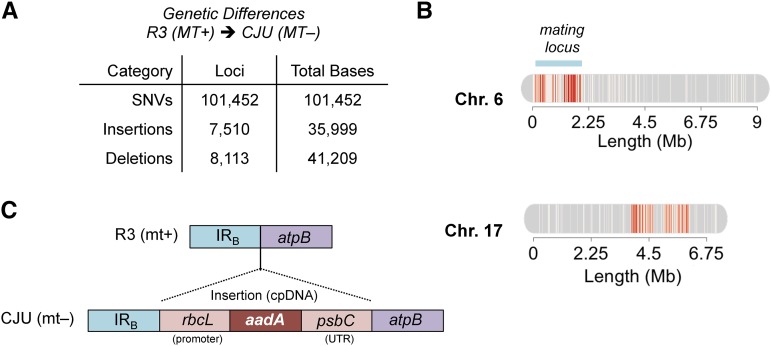
As a prelude to transcriptome and methylome analyses, we cataloged the genetic differences (single-nucleotide variants, insertions, and deletions) between our two parental strains using genome resequencing (Fig. 2A; Gallaher et al., 2015). In total, the two strains differ by 0.16% in their nuclear genomes, and most of these differences are single-nucleotide variants. Of all the variants, 98.2% are localized to two regions, one of length 2.2 Mb on chromosome 17 and one of 2 Mb encompassing the mating locus on chromosome 6 (Fig. 2B), where mating-type haplotype differences have been observed previously (De Hoff et al., 2013). Additionally, the chloroplast genome of the CJU10 strain contains an insertion adjacent to the ATP synthase subunit betaaadA (atpB) gene of a spectinomycin resistance marker (aminoglycoside-3′-adenylyltransferase [aadA]) flanked by the Rubisco large subunit (rbcL) 5′ promoter and photosystem II CP43 (psbC) 3′ untranslated region (Fig. 2C; Goldschmidt-Clermont, 1991; Umen and Goodenough, 2001).
Figure 2.
Genetic differences between R3 (MT+) and CJU10 (MT−) parental strains. A, Genetic differences, categorized by variant type (single-nucleotide variants [SNVs], insertions, and deletions), are shown with the total number of variant loci and total bases. B, Large-scale view of chromosomes 6 and 17, where 98.5% of the identified variants are located. Locations of sequence differences are shown in red. The region of chromosome 6 containing the mating locus is denoted by the blue bar. C, Schematic representation of the antibiotic resistance transgene insertion in CJU10 cpDNA. The aadA gene, along with the rbcL 5′ promoter and the psbC 3′ untranslated region (UTR), is inserted between inverted repeat region B (IRB) and the endogenous atpB gene.
Gamete and Zygote-Specific Genes
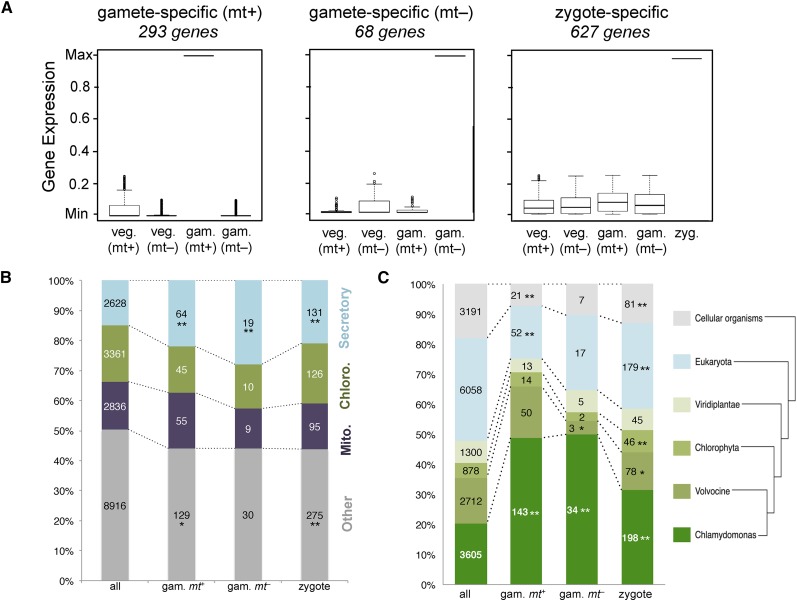
Following the quantitation of RNA-seq data for all of our samples, we identified genes with mating type- and zygote-specific expression patterns using a series of filters to screen for genes matching the expression patterns of known gametic and zygotic genes. We required that mating type-specific genes be expressed in gametes at least 4-fold higher than in vegetative cells of the same mating type and at least 10-fold higher than in gametes or vegetative cells of the opposite mating type. For zygote-specific genes, we required that expression be at least 4-fold higher in early zygotes than in any other sample. Using these criteria, we identified 293 and 68 genes whose expression is specific to plus and minus gametes, respectively, and 627 genes whose expression is specific to zygotes (Fig. 3A; Supplemental Table S1). Genes whose expression is known to be gamete or zygote specific, including GSM1 (minus gametes), SAG1 (plus gametes), and EZY1 (early zygotes), were retained by our filters (Armbrust et al., 1993; Kurvari et al., 1998; Ferris et al., 2002, 2005; Lee et al., 2008). Of the 32 previously described zygotic genes (Matters and Goodenough, 1992; Armbrust et al., 1993; Uchida et al., 1993, 1999; Kuriyama et al., 1999; Suzuki et al., 2000; Ferris et al., 2002; Kubo et al., 2008), 25 were in our zygote data set, while the remaining seven (EZY6, EZY15, EZY16, EZY17, EZY21, EZY23, and Lysozome 1A) were found to have significant expression in other samples and, therefore, were excluded from the zygote-specific set (Supplemental Tables S1 and S2).
Figure 3.
Gamete- and zygote-specific genes. A, Box and whisker plots of gene expression profiles for gamete- and zygote-specific genes. Values are plotted relative to the maximum expression value of each of the genes. A total of 293 and 68 genes were expressed specifically in MT+ and MT– gametes, respectively. A total of 627 genes were expressed specifically in zygotes. B, Localization predictions for proteins encoded by gamete-specific (gam. mt+ and gam. mt−) and zygote-specific genes, compared with all predicted proteins in the C. reinhardtii proteome (all). Data are plotted as the fraction predicted for each of four compartments, with total numbers indicated within each graph portion: secretory in blue; chloroplast (Chloro.) in green; mitochondria (Mito.) in purple; and other in gray. Values with asterisks are significantly different from the total proteome distribution (*, P < 0.05 and **, P < 0.01). C, Predicted protein groups as described in B are plotted according to the taxonomic distribution of encoded proteins from each category. From bottom (C. reinhardtii-specific proteins; dark green) to top (all cellular organisms; gray) are increasingly broad phylogenetic distributions. Samples with asterisks indicate significant enrichment or depletion in a taxonomic category relative to the distribution of all proteins (*, P < 0.05; and **, P < 0.01).
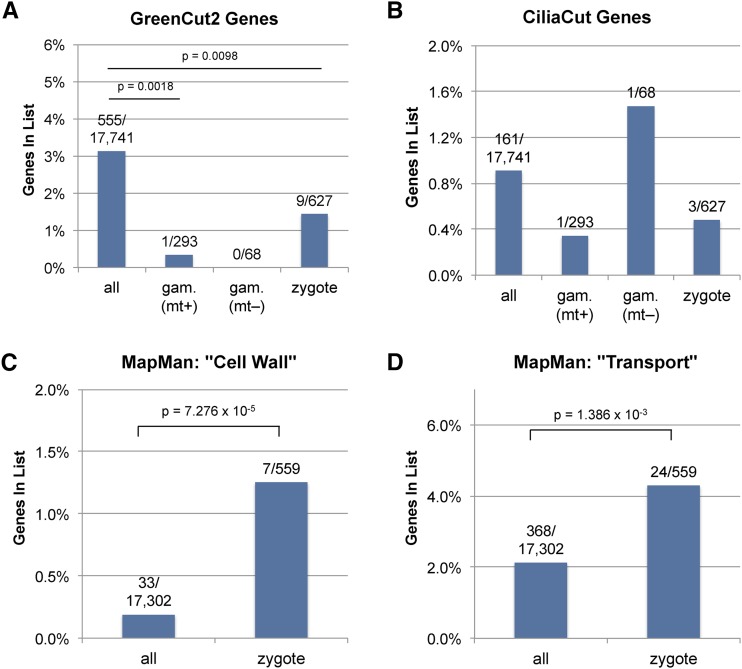
We functionally classified the predicted proteins encoded by genes whose expression was plus gamete specific, minus gamete specific, or early zygote specific. Protein localization prediction revealed the enrichment of putative secretory proteins in plus-gamete (MT+) and zygote-specific sets (Fig. 3B). We also assessed the conservation and phylogenetic distribution for homologs of each protein in a set of nested phylogenetic domains that encompass different taxonomic levels from cellular organisms (prokarya, archaea, and eukarya) to the single species level of C. reinhardtii (Fig. 3C). Zygote and gamete expression groups were enriched for C. reinhardtii-specific genes and/or alga-specific genes. In addition, gametic genes were underrepresented for more widely conserved genes (i.e. those with homologs outside of chlorophyte algae). Consistent with these findings, the gametic and zygotic gene lists were also depleted to various extents for GreenCut2 and CiliaCut genes, whose members are associated with conserved photosynthetic and flagella/basal body functions (Merchant et al., 2007; Karpowicz et al., 2011; Heinnickel and Grossman, 2013; Fig. 4, A and B); however, the overall low numbers of genes in these categories precluded obtaining a significant statistical result in all cases but one. Gene Ontology, Kyoto Encyclopedia of Genes and Genomes, and MapMan (Kanehisa and Goto, 2000; Harris et al., 2004; Thimm et al., 2004) classification of predicted proteins encoded by gametic and zygotic genes was performed using the Algal Functional Annotation Tool (Lopez et al., 2011) but had limited utility because of the large number of nonconserved proteins in these groups and incomplete annotations. Nonetheless, we found significant enrichment in two MapMan categories for zygotic genes, cell wall and transport, both of which may relate to the requirement for new cell wall biosynthesis in zygotes (Fig. 4, C and D). Indeed, examination of manually curated early zygotic gene annotations revealed numerous cell wall-related protein-coding genes as described below.
Figure 4.
Functional annotation of proteins encoded by gamete- and zygote-specific genes. Functional annotations for gamete- and zygote-specific gene lists are shown as bar plots showing the fraction of total proteins in each group with the specified annotation. The total number of genes in each annotation category versus the number of genes with the described annotation are shown. Significant differences between all genes and gamete- or zygote-specific genes were calculated with the hypergeometric test, and significant P values are shown. A, GreenCut2 genes (Karpowicz et al., 2011; Heinnickel and Grossman, 2013). B, CiliaCut genes (Merchant et al., 2007). C, MapMan cell wall-related genes (Thimm et al., 2004). D, MapMan transport-related genes.
Volvocine cell walls are composed primarily of glycosylated hydroxyproline-rich glycoproteins (HRGPs) that enter the secretory pathway and are exported to the extracellular space where they coassemble (Woessner et al., 1994). The thick and environment-resistant cell walls of zygospores are formed by a specialized set of HRGPs that are synthesized shortly after fertilization (Minami and Goodenough, 1978; Catt, 1979; Grief et al., 1987). Manual annotation and inspection of zygotic up-regulated genes verified the MapMan ontology assignments of cell wall and transport categories as described in Supplemental Table S1. At least 57 zygotic genes are predicted HRGPs or have putative cell wall biogenesis-related functions that include secretion, glycosylation, and metabolism of nucleotide sugars (e.g. UDP-Glc 4-epimerase, pyrophosphorylase, dehydrogenase, dTDP-6-deoxy-l-lyxo-4-hexulose reductase related, and exotosin-like glycosyltransferase) or sugar metabolite transport (e.g. ATP-binding cassette transporter, triose phosphate transporter, and UDP-GlcNAc transporter). Among these were some previously identified early zygotic genes as noted in Supplemental Table S1 (EZY4, EZY11/UDP-glucose:protein transglucosylase1 [UPT1; also known as UPTG1]/EZY12/UDP-glucose dehydrogenase1, EZY14/triose phosphate transporter14, and EZY22; Kubo et al., 2008).
Besides cell wall and secretory pathway genes, we also noted in the zygote gene set predicted functions that may be related to other zygotic processes, including elimination of MT– cpDNA, zygotic cpDNA methylation, and packaging for long-term dormancy, nuclear fusion (karyogamy), chloroplast fusion, and flagellar resorption (Goodenough et al., 2007). These annotations include predicted chloroplast-targeted DNA-binding proteins such as a DNA recombination protein A homolog and predicted nuclease (EZY19/Cre07.g314650), chloroplast-targeted DNA methyltransferases (DMT1A, DMT1B, and DMT4; discussed below), and a chloroplast-targeted dynamin (EZY8/Cre06.g25065) that may be involved in chloroplast fusion (Kubo et al., 2008). Predicted nucleus-targeted zygotic proteins include several DNA-binding transcription factors (EZY18/Cre02.g091550, Regeneration Protein A [RegA]/RlsA-like protein7/Cre14.g617200, and zygote-specific transcription factor 1A/Cre17.g719200) such as RLS7 that contains a SAND domain (Duncan et al., 2006, 2007) related to RegA, a repressor of germ cell fate in Volvox carteri (Kirk et al., 1999), and several types of chromatin-related proteins (Cre03.g184900, Cre08.g367000, Cre08.g400200, Cre09.g401812, and histone H1/Cre13.g567450) that may be involved in nuclear DNA packaging in preparation for zygospore dormancy. Minutes after fertilization and prior to flagellar resorption, the four basal bodies and flagella of newly formed zygotes (two from each parent) move to a single apical location via an unknown mechanism. One possible participant in this process could be the striated fiber protein SF-assemblin (Cre07.g332950), a zygote up-regulated gene whose protein product in vegetative cells associates with rootlet microtubules that are proximal to basal bodies and are thought to play a role in the organization of the rootlet structure (Lechtreck et al., 2002). Two other cytoskeletal proteins whose genes are up-regulated in zygotes are a flagella-associated protein of unknown function, FAP79 (Cre04.g217908), and the flagella length regulatory protein LF5/FAP279 (Cre12.g538300; Tam et al., 2013), which may be related to flagellar resorption that begins 2 or 3 h after fertilization. Lastly, we identified a gene for a predicted secreted trypsin-related protease (Cre06.g287750), which could contribute to the rapid postfertilization degradation of gametic plasma membrane surface proteins such as fusion protein1 and generative cell specific1, a process that is thought to restrict polygamy (fusion between more than two gametes; Liu et al., 2010).
DNA Methylation of the Nuclear Genome
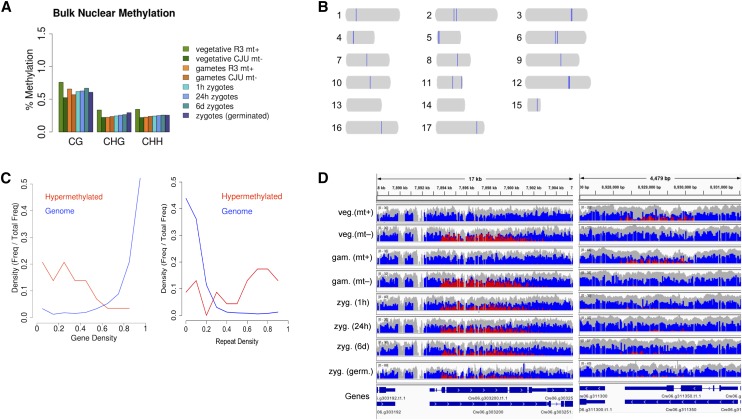
We conducted bisulfite sequencing of vegetative, gametic, and zygotic samples to generate DNA methylation profiles. The nuclear genome had an average per-site CG methylation of less than 0.75% in all samples, and this level of methylation did not differ significantly between plus and minus strains or at different life cycle stages (Fig. 5A). However, CG methylation densities greater than 80% were identified for 23 loci that ranged in size between 10 and 22 kb (Fig. 5B; Supplemental Table S3). The highly methylated regions are enriched for repeats, and their overall protein-coding gene densities are significantly lower than average, although there are still genes in these regions (Fig. 5C). In addition, one example where methylation was strain specific is shown in Figure 5D, although most hypermethylated sites did not have strain-specific methylation patterns. The expression levels of genes overlapping hypermethylated loci are not strongly correlated with degree of methylation (Supplemental Table S4).
Figure 5.
Nuclear methylation at different C. reinhardtii life cycle stages. A, Bulk averages of 5meC in the nuclear genome for each sample and categorized by the fraction of 5meC in each of three sequence contexts (CG, CHG, and CHH). B, Scaled representations of the 17 C. reinhardtii chromosomes, with 23 hypermethylated regions larger than 10 kb shaded in blue. C, Plots comparing gene and repeat densities of hypermethylated regions relative to the entire genome. D, Genome browser display of nuclear cytosine methylation frequency at two representative hypermethylated loci from chromosome 6. Two representative examples of strain-specific hypermethylated loci are shown side by side, with a gene track below for genomic context. In the coverage plot, blue represents unmethylated cytosines (converted by bisulfite treatment), red represents methylated cytosines (unconverted), and gray represents total coverage (including reads from the opposite strand that do not provide methylation information).
Chloroplast Methylation Changes during the Life Cycle
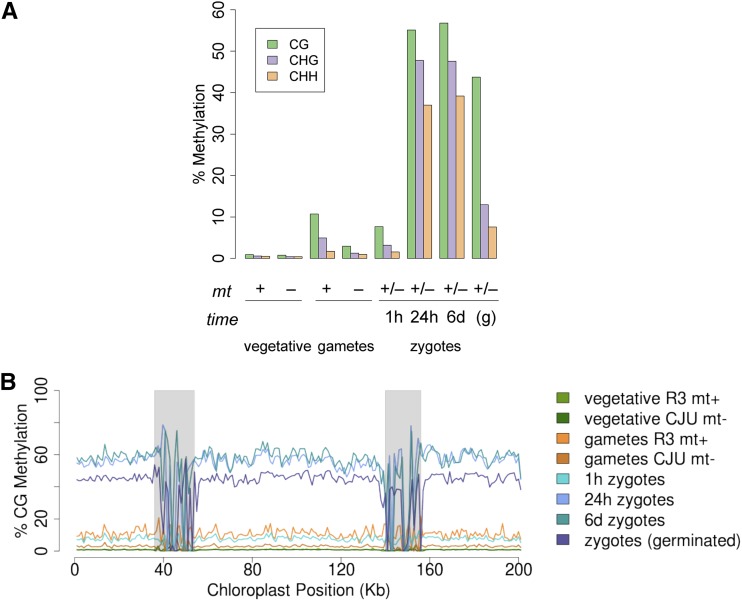
In contrast to the relatively stable pattern of cytosine methylation in nuclear DNA, the C. reinhardtii chloroplast genome underwent dynamic changes in cytosine methylation throughout the life cycle (Fig. 6A). In the vegetative stage, global per-cytosine methylation was less than 2% for both mating types for all cytosine contexts (CG, CHG, and CHH). After gametogenesis, cpDNA methylation increased in a mating type-dependent manner. MT+ gametes had an average of approximately 10% per-site CG methylation, while MT− gametes had an average of approximately 3%. A large increase in 5meC was observed for all sequence contexts during zygote development, with 54% (CG) methylation reached by 24 h. Methylation levels began to drop during germination to an average per-site CG methylation level of 45%. CHG and CHH methylation levels were lower than CG methylation levels in all cases, and this difference was particularly notable in germinated zygotes, where CHH and CHG methylation dropped from their peak zygotic levels much faster than CG methylation did.
Figure 6.
Chloroplast genome methylation at different life cycle stages. A, Bulk cytosine methylation frequencies for cpDNA at different life cycle stages plotted for each methylation context. The mating type of each sample (+, −, or diploid +/−) and time of zygote development and germination (g) are shown below the graph. B, Plot of average CG methylation frequency for each sample in 1-kb bins across the chloroplast genome. The two inverted repeat regions are shaded in gray. Plots are color coded according to the legend at right.
For all life cycle stages and mating types, methylation of cpDNA is uniformly distributed in the chloroplast genome without bias toward genes or other sequence features (Fig. 6B). With the exception of two large inverted repeats in the chloroplast sequence, where methylation cannot be measured accurately with short reads, average methylation levels tabulated in 1-kb intervals rarely deviated more than 5% from the global average, suggesting that no specific regions of the chloroplast genome are targeted for methylation.
DNA Methyltransferases
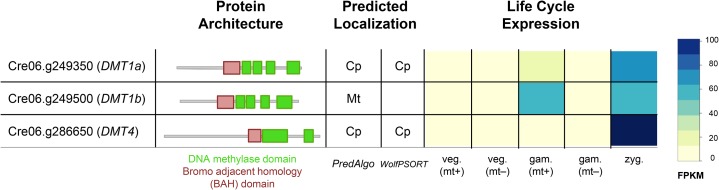
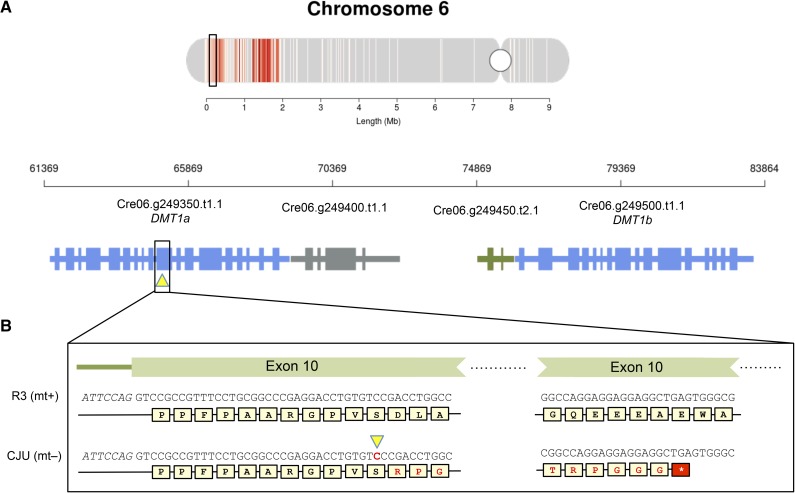
In order to further explore the mechanisms responsible for the dynamic patterns of DNA methylation, we identified candidate cytosine methyltransferases from the predicted C. reinhardtii proteome based on the presence of predicted DNA cytosine methylase domains (see “Materials and Methods”). We found a total of six candidate methyltransferases with different domain architectures (Supplemental Fig. S1), including a homolog of V. carteri MET1, a putative nuclear methyltransferase (Babinger et al., 2007). As described below, three predicted methyltransferases, DMT1a, DMT1b, and DMT4, had expression patterns and/or predicted localization sequences suggesting a role in cpDNA methylation. Each of these predicted proteins has DNA methylase as well as bromo-adjacent homology (BAH) domains. DMT1a, DMT1b, and DMT4 were all expressed in gametes and showed the highest levels of expression in zygotes, coinciding with elevated levels of methylation in the zygotic samples (Fig. 7). DMT1a and DMT1b are near the mating locus in its telomere-proximal domain and encode highly similar paralogs. Both genes had higher expression in MT+ than in MT− gametes, a pattern that matches the methylation bias seen in gametic cpDNA from the two mating types (Fig. 6A). DMT1a and DMT1b sequences have been described previously as a single gene (Nishiyama et al., 2002), but the published cDNA sequence is a hybrid with 5′ sequences derived from DMT1a and 3′ sequences from DMT1b (National Center for Biotechnology Information [NCBI] gene identifiers 5722229 and 5722231). Consistent with subcellular targeting predictions (Fig. 7), the DMT1a presequence directs chloroplast localization (Nishiyama et al., 2002). While the MT+ strain copy of DMT1a appears to be intact, a survey of structural variants derived from genome resequencing data led to the identification of a point insertion in exon 10 of the MT− strain copy of DMT1a leading to a frame shift and premature termination before the methyltransferase domains (Fig. 8B). This insertion was observed in MT– transcriptome data. Furthermore, this point insertion is found in the genome of 12 out of 13 MT– strains resequenced by Gallaher et al. (2015). No variants predicted to be deleterious were found within the DMT1b gene. Targeting predictions of DMT1b suggest that it is mitochondria localized. Although the mitochondrial genome is largely devoid of cytosine methylation (1%–2% global per-cytosine methylation), the zygote sample at 24 h shows evidence of methylation (approximately 13% global per-cytosine methylation; Supplemental Fig. S2). DMT4 is also predicted to encode a chloroplast-targeted cytosine methyltransferase and is one of the 361 genes identified with a strong zygotic expression pattern (Fig. 3A), consistent with a possible participation of DMT4 in zygotic cpDNA hypermethylation.
Figure 7.
Candidate chloroplast methyltransferases in C. reinhardtii. The protein domain structure of each candidate methyltransferase is shown schematically (green = DNA methylase domain and red = BAH domain). Predicted localizations from PredAlgo and WolfPSORT (Cp = chloroplast and Mt = mitochondria) are shown alongside the log-transformed normalized expression level of each candidate from different RNA-seq samples. FPKM, Fragments per kilobase of transcript per million mapped reads.
Figure 8.
Single-base insertion variant in DMT1a leads to a premature stop codon. A, The DMT1 locus on chromosome 6 containing DMT1a and DMT1b is shown below its context in the entire chromosome. B, The diagram at top shows an enlarged chromosomal region containing the DMT1 locus with genomic coordinates and adjacent genes. The position of the insertion variant in DMT1a is indicated by the yellow triangle. Below is an expanded view of exon 10 from DMT1a showing the single-base insertion in the MT– strain (CJU10) and the altered protein-coding sequence and premature stop codon caused by the cytosine insertion, shown in boldface and marked with the yellow triangle.
DISCUSSION
The dynamic changes in gamete- and zygote-specific mRNA abundance and DNA methylation presented in this work provide a framework for understanding cell differentiation during the C. reinhardtii sexual life cycle. A previous study of plus and minus gamete-specific genes focused on cell type genes whose expression was up-regulated during the process of mating (Ning et al., 2013). However, direct comparison with the previous data is confounded by differences in annotation between genome versions used to define transcripts as well as in the different clustering criteria used in the two studies. Here, we made use of culture conditions that were designed to suppress the differential transcript abundance signal resulting from growing versus nongrowing nitrogen-starved cultures to help identify gamete-specific transcripts. Our method identified known gametic genes whose expression is mating type limited (expressed preferentially in plus versus minus gametes or vice versa). However, because we used nitrogen resupply of stationary phase cultures to create nongrowing vegetative samples, we may have missed highly stable gametic transcripts that did not turn over after nitrogen addition. Nonetheless, nitrogen resupply for several hours was completely effective at suppressing mating, so the transcriptome differences we were able to identify in our gametic versus vegetative samples are likely tied to gametogenesis-related functions and perhaps less relevant for other nitrogen-starvation responses such as neutral lipid accumulation.
Sex-related genes evolve rapidly and, therefore, are expected to appear younger and have a more restricted phylogenetic distribution than other genes (Swanson and Vacquier, 2002). Indeed, using phylogenomic profiling, we found an enrichment of C. reinhardtii-specific genes belonging to the plus or minus gamete up-regulated categories and zygote up-regulated category (Fig. 3C). This finding underscores the importance of species-specific and clade-specific genes as potential drivers of cell type specialization related to sex and speciation. Although the functions of these genes are difficult to predict, since they have no homologs outside of C. reinhardtii or volvocine algae, we did find enrichment for secretory pathway targeting signals within the plus gamete and zygote predicted proteins (Fig. 3B), which could indicate a role for sex-related proteins in the plasma membrane, cell wall, or extracellular space. Manual annotation of zygotic genes showed that their predicted functions match processes such as glycosylation and transport that are associated with cell wall formation. In addition, we identified or confirmed the expression of zygotic genes that may be associated with other poorly understood differentiation processes, including chloroplast fusion, MT– cpDNA elimination, DNA methylation, and cytoskeletal remodeling (Supplemental Table S1). Deeper investigation of these genes may yield insights into the cell biology of zygote differentiation that may have parallels in other zygospore-forming algae (Brawley and Johnson, 1992) and even in plants where pollen cells must undergo a similar process of dormancy and reactivation when exposed to appropriate conditions (Brown and Lemmon, 2011). Early zygotes in C. reinhardtii also undergo dramatic changes in their cytoskeleton. Unlike the case in animals, where paternal but not maternal centrioles are contributed to zygotes (Avidor-Reiss et al., 2015), in C. reinhardtii, both parental basal bodies (structurally similar to centrioles) and flagella are retained initially in zygotes but eventually are disassembled and rebuilt during germination (Cavalier-Smith, 1976). Nonetheless, investigation of conserved zygotically up-regulated cytoskeletal proteins such as LF5 and SF-assemblin may shed light on more general mechanisms involved in controlling the dynamic behavior of flagella/cilia and basal bodies during cellular remodeling and life cycle transitions.
DNA Methylation Changes during the Life Cycle
In contrast to many organisms in which a large fraction of the nuclear genome is methylated (Smith and Meissner, 2013; Bestor et al., 2015), we found relatively low levels of cytosine methylation in the nuclear genome of C. reinhardtii, consistent with previous surveys that were not as high resolution as reported here (Hattman et al., 1978; Feng et al., 2010). Interestingly, we did identify 23 hypermethylated loci larger than 10 kb (Supplemental Table S3) that tended to occur within gene-poor, repeat-rich regions of the genome. However, the mechanism that leads to the methylation of these loci remains unknown. Previous studies on the silencing of transgenes in C. reinhardtii found that inserted transgene repeats were frequently methylated, but they reported little correlation between methylation and gene expression (Cerutti et al., 1997). On the other hand, in the related colonial alga V. carteri, where methylcytosine frequency is slightly higher than in C. reinhardtii (1.1% versus approximately 0.75%), nuclear methylation did appear to be associated with gene silencing (Babinger et al., 2001, 2007). Interestingly, V. carteri also has a more repeat-rich genome than C. reinhardtii with many active transposons (Miller et al., 1993; Ueki and Nishii, 2008; Prochnik et al., 2010) and may have retained or evolved higher levels of nuclear methylation activity than C. reinhardtii to suppress transposon activity.
The low frequency of cytosine methylation in the nuclear genome contrasts with the dynamic and abundant cytosine methylation in the chloroplast genome. The overall patterns we observed are in agreement with previous findings that vegetative cpDNA from both mating types had low levels of cytosine methylation and that gametes showed elevated levels, with MT+ cpDNA having a 2- or 3-fold higher frequency of methylcytosine compared with MT– cpDNA (Royer and Sager, 1979; Dyer, 1982). We observed hypermethylation in zygotes, which has also been seen previously. Our study extended these earlier results by examining genome-wide methylation in cpDNA at single-base resolution. Unlike the methylation of nuclear DNA, which was mostly restricted to a few loci, cpDNA cytosine methylation was uniformly distributed across all regions and sequence feature types (genic, intergenic, repeats, exons, introns, etc.). This finding has implications for the function of cpDNA methylation and the enzymes that are responsible for methylating cpDNA (elaborated in the next section). Previous models of cpDNA inheritance invoked a methylation-restriction system similar to that in prokaryotes where sequence-specific methyltransferases protect MT+ cpDNA from site-specific restriction endonucleases. However, the non-sequence-specific distribution of methylcytosines we observed and the modest differences between levels of MT+ and MT– cpDNA methylation in gametes are not consistent with a methylation-restriction mechanism. Moreover, if cpDNA methylation were related to methylation and restriction, it would be most effective if it were established at the gametic stage of the life cycle. Instead, the majority of cpDNA methylation occurs in zygotes and is pervasive genome wide. The purpose of this massive methylation is unknown but is likely tied to the packaging and protection of cpDNA in zygospores, where it may be dormant for many years before germination (Brawley and Johnson, 1992).
DNA Methyltransferases
Our survey of candidate methyltransferases revealed two candidates with expression patterns and predicted chloroplast targeting sequences that make them likely responsible for cpDNA methylation: DMT1a and DMT4. Both genes have detectable expression in gametes and zygotes (Fig. 7). DMT1a and DMT1b are physically and genetically linked to the mating locus and, furthermore, the MT– linked copy of DMT1a has a point mutation that is predicted to generate a truncated, nonfunctional protein. Although they are linked to the mating locus, the DMT1a/b locus could still recombine with the mating-type locus or be subject to gene conversion, possibly leading to the creation of strains where the levels of gametic cpDNA in MT+ and MT– parents are equivalent. Such strains, if they could be isolated, would be useful for testing the significance of differential gametic cpDNA methylation and the contribution of the DMT1a gene to gametic cpDNA methylation levels.
Although the predicted DMT1- and DMT4-encoded methyltransferases have accessory BAH domains that are implicated in protein-protein interactions and targeting to specific loci (Callebaut et al., 1999; Yang and Xu, 2013), the nonspecific pattern of cpDNA methylation we observed and attribute to the predicted DMT1a and DMT4 proteins suggests that their targeting is not sequence specific. The BAH domains in these proteins may serve other functions, such as general targeting of the methyltransferase enzymes to chloroplast nucleoids. The lack of sequence specificity predicted for DMT1a and DMT4 may also prove useful for biotechnology applications that require nonspecific DNA cytosine methylation activity.
MATERIALS AND METHODS
Sample Generation for Bisulfite-Treated DNA
Chlamydomonas reinhardtii strains R3 (CC-620 R3 NM MT+) and CJU10 (Umen and Goodenough, 2001) were grown in high-salt medium to concentrations of 3.4 × 106 and 3.5 × 106 cells mL−1, respectively. A total of 100 mL of each strain culture was used for DNA and RNA isolation as vegetative samples. Cells were collected by centrifugation (3,500 rpm for 5 min), resuspended in high-salt medium without nitrogen, and after 15 h, 40 mL of each strain culture was collected as gametic samples. Gametes were mixed and checked for mating efficiency (85% efficient), and after 1 h, a 40-mL sample was collected, corresponding to the 1-h zygote sample. Mixed gametes were split into two flasks: (1) cells in the first were collected after 24 h, corresponding to the 24-h zygote sample; (2) cells in the second were resuspended in water and plated with high-salt medium, incubated in the light for 24 h, incubated for 5 d in the dark, and resuspended in Tris-EDTA-NaCl (TEN) buffer with 0.2% (v/v) Nonidet P-40, at which time half of the culture was collected as 6-d zygote samples. The remaining half was transferred to Tris-acetate phosphate medium, incubated in the light for 24 h, and collected as the germinated zygote sample.
DNA Isolation and Purification for Bisulfite Treatment
Cells were resuspended in 4 mL of TEN buffer, with the exception of 24-h and 6-d zygotes. The 24-h and 6-d zygotes were resuspended in 10 mL of TEN, mixed for 1 min, and repelleted (repeated three times) followed by resuspension in 4 mL of TEN. A total of 400 μL of 20% (w/v) SDS and 400 μL of 20% (v/v) sarkosyl was added to each sample. For 24-h and 6-d zytores, 4 mL of zirconium beads was also added followed by vortexing for 5 min. A total of 200 μL of pronase E solution (10 mg mL−1) was then added, and samples were incubated at 37°C for 30 min. A total of 2.5 mL of phenol (10 mm Tris-Cl, pH 8, saturated) was added, followed by 2.5 mL of chloroform:isoamylalcohol (24:1). The phases were separated by centrifugation (5,000 rpm for 10 min) at 10°C, and the upper phase (4.5 mL) was transferred into 9 mL of 100% ethanol and incubated overnight at −20°C. Nucleic acids were collected by centrifugation, resuspended in 1 mL of 10 mm Tris-Cl, pH 8, and 100 μL of RNase (5 mg mL−1) was added. After RNase treatment, samples were extracted again with 1 mL of phenol:chloroform:isoamylalcohol (25:24:1), and DNA was precipitated with 0.3 m sodium acetate and 70% (v/v) isopropanol for 30 min at room temperature.
DNA Library Preparation
A total of 500 ng of purified genomic DNA was sheared by sonication with Covaris S2 to generate DNA fragments spanning from 100- to 400-bp size range. Library preparation was carried out using NEBNext DNA Library Prep Master Mix (Set for Illumina; New England Biolabs; catalog no. E6040) according to the manufacturer’s instructions with minor modifications. The ligation was performed using Illumina TruSeq Adapters (catalog no. 15025064), and DNA size selection (200- to 400-bp range) was carried out with AMPure XP beads (Beckman Coulter) prior to bisulfite conversion using the EZ DNA Methylation-Lightning Kit (Zymo; catalog no. D5030). The bisulfite-treated DNA was amplified using Illumina Primer Cocktail Mix (catalog no. 15027084) and MyTaq Mix (Bioline; catalog no. BIO-25045) according to the following program: 98°C for 2 min; 12 cycles of 98°C for 15 s, 60°C for 30 s, and 72°C for 30 s; and then 72°C for 5 min.
RNA Library Preparation
Total RNA isolation for RNA-seq analysis was performed as described previously (De Hoff et al., 2013) with additional DNase treatment (2 units of Roche RNase-free DNase per 110 μg of total RNA, 37°C for 20 min) before final Qiagen RNeasy column purification.
Genomic Variation
Genomic reads were aligned using Burrows-Wheeler Aligner version 0.6.2 (Li and Durbin, 2010), with default parameters, to the version 5.0 assembly of the C. reinhardtii CC-503 genome (Merchant et al., 2007). After removing duplicates with Picard MarkDuplicates (http://broadinstitute.github.io/picard/), we applied Genome Analyzer Tool Kit (McKenna et al., 2010) base quality score recalibration, insertion/deletion realignment, and small variant discovery (DePristo et al., 2011). This was followed by hard filtering of variants with extensive manual calibration guided by inspections in Integrative Genomics Viewer (Thorvaldsdóttir et al., 2013).
RNA-seq Analysis
RNA-seq data were aligned to the C. reinhardtii genome (assembly version 5.5; Phytozome version 10.3 gene annotation) with TopHat version 2.0.10 using the annotation to guide spliced alignment. Default parameters were kept, with the exception of constraining intron lengths to less than 5 kb. Expression levels were quantified using Cufflinks version 2.2.1 to compute fragments per kilobase of transcript per million mapped reads.
Identification of Gamete- and Zygote-Specific Genes
Gamete- and zygote-specific genes were identified by applying a series of filters to the fragments per kilobase of transcript per million mapped reads data generated as described above. We defined gamete-specific genes for each mating type as those that have expression in all samples (excluding the zygote) that is less than 10% of the expression level in the gamete of that mating type. Zygote-specific genes were defined as those whose expression in all other samples was less than 25% of the expression level in the zygote. Expression values for samples with replicates were averaged.
Bisulfite-Treated DNA Sequencing Analysis
Raw sequence data were demultiplexed using standard Illumina barcode indices and checked for quality using FastQC (version 0.10.1). Bisulfite-converted sequences were aligned to the C. reinhardtii nuclear genome (November 2011 assembly) and chloroplast genome using the BS-Seeker2 alignment pipeline version 2.0.5 (Guo et al., 2013). Default whole-genome bisulfite alignment parameters were chosen with the following exceptions: Bowtie2 was used as the aligner, and local alignments were enabled. Methylation levels were called for cytosines covered by at least four reads. Sequence data have been deposited in NCBI’s Short Read Archive under the accession numbers SRR2051057, SRR2051058, SRR2051059, SRR2051060, SRR2051061, SRR2051062, SRR2051063, and SRR2051065.
Identification of Candidate Methyltransferases
To identify candidate DNA methyltransferases, protein sequences of C. reinhardtii (Phytozome version 10 gene annotation) were scanned against the Pfam-A database (release 27) using the stand-alone PfamScan scripts provided by the Wellcome Trust Sanger Institute. The presence of the C-5 cytosine-specific DNA methylase domain (PF00145) was used as a criterion for assignment as a cytosine methyltransferase.
Sequence data have been deposited in NCBI's Short Read Archive under the accession numbers SRR2051057, SRR2051058, SRR2051059, SRR2051060, SRR2051061, SRR2051062, SRR2051063, and SRR2051065.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Candidate methyltransferases in C. reinhardtii.
Supplemental Figure S2. Methylation patterns for C. reinhardtii mitochondrial DNA.
Supplemental Table S1. Annotation of gamete and zygote specific genes.
Supplemental Table S2. Expression values for gamete and zygote specific genes.
Supplemental Table S3. Coordinates of hypermethylated regions in nuclear genome with repeat data.
Supplemental Table S4. Methylation and expression data for genes in regions with strain specific methylation patterns.
Supplementary Material
Glossary
- cpDNA
chloroplast DNA
- 5meC
5-methylcytosine
- RNA-seq
RNA sequencing
- HRGPs
hydroxyproline-rich glycoproteins
- BAH
bromo-adjacent homology
- NCBI
National Center for Biotechnology Information
- TEN
Tris-EDTA-NaCl
Footnotes
This work was supported by the National Institutes of Health (grant nos. R24 GM092473 and R01 GM078376 and T32 Training Fellowship in Genome Analysis no. 5T32HG002536–13 to D.L.); by the Office of Science (Biological and Environmental Research), U.S. Department of Energy (grant no. DE–FC02–02ER63421); by the Eugene V. Cota-Robles Fellowship and the Fred Eiserling and Judith Lengyel Doctoral Fellowship (to D.L.); and by the Japan Society for the Promotion of Science (Postdoctoral Fellowship for Research Abroad no. 26–495 to T.H.).
References
- Albee AJ, Kwan AL, Lin H, Granas D, Stormo GD, Dutcher SK (2013) Identification of cilia genes that affect cell-cycle progression using whole-genome transcriptome analysis in Chlamydomonas reinhardtii. G3 (Bethesda) 3: 979–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama H, Saitoh S, Kuroiwa T, Nakamura S (2014) Comparative analysis of zygospore transcripts during early germination in Chlamydomonas reinhardtii. J Plant Physiol 171: 1685–1692 [DOI] [PubMed] [Google Scholar]
- Armbrust EV, Ferris PJ, Goodenough UW (1993) A mating type-linked gene cluster expressed in Chlamydomonas zygotes participates in the uniparental inheritance of the chloroplast genome. Cell 74: 801–811 [DOI] [PubMed] [Google Scholar]
- Avidor-Reiss T, Khire A, Fishman EL, Jo KH (2015) Atypical centrioles during sexual reproduction. Front Cell Dev Biol 3: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babinger P, Kobl I, Mages W, Schmitt R (2001) A link between DNA methylation and epigenetic silencing in transgenic Volvox carteri. Nucleic Acids Res 29: 1261–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babinger P, Völkl R, Cakstina I, Maftei A, Schmitt R (2007) Maintenance DNA methyltransferase (Met1) and silencing of CpG-methylated foreign DNA in Volvox carteri. Plant Mol Biol 63: 325–336 [DOI] [PubMed] [Google Scholar]
- Bestor TH, Edwards JR, Boulard M (2015) Notes on the role of dynamic DNA methylation in mammalian development. Proc Natl Acad Sci USA 112: 6796–6799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaby IK, Blaby-Haas CE, Tourasse N, Hom EF, Lopez D, Aksoy M, Grossman A, Umen J, Dutcher S, Porter M, et al. (2014) The Chlamydomonas genome project: a decade on. Trends Plant Sci 19: 672–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolen PL, Grant DM, Swinton D, Boynton JE, Gillham NW (1982) Extensive methylation of chloroplast DNA by a nuclear gene mutation does not affect chloroplast gene transmission in Chlamydomonas. Cell 28: 335–343 [DOI] [PubMed] [Google Scholar]
- Boyle NR, Page MD, Liu B, Blaby IK, Casero D, Kropat J, Cokus SJ, Hong-Hermesdorf A, Shaw J, Karpowicz SJ, et al. (2012) Three acyltransferases and nitrogen-responsive regulator are implicated in nitrogen starvation-induced triacylglycerol accumulation in Chlamydomonas. J Biol Chem 287: 15811–15825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawley SH, Johnson LE (1992) Gametogenesis, gametes and zygotes: an ecological perspective on sexual reproduction in the algae. Br Phycol J 27: 233–252 [Google Scholar]
- Brown RC, Lemmon BE (2011) Spores before sporophytes: hypothesizing the origin of sporogenesis at the algal-plant transition. New Phytol 190: 875–881 [DOI] [PubMed] [Google Scholar]
- Burton WG, Grabowy CT, Sager R (1979) Role of methylation in the modification and restriction of chloroplast DNA in Chlamydomonas. Proc Natl Acad Sci USA 76: 1390–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callebaut I, Courvalin JC, Mornon JP (1999) The BAH (bromo-adjacent homology) domain: a link between DNA methylation, replication and transcriptional regulation. FEBS Lett 446: 189–193 [DOI] [PubMed] [Google Scholar]
- Catt JW. (1979) Isolation and chemical composition of the zygospore cell wall of Chlamydomonas reinhardtii. Plant Sci Lett 15: 69–74 [Google Scholar]
- Cavalier-Smith T. (1976) Electron microscopy of zygospore formation in Chlamydomonas reinhardtii. Protoplasma 87: 297–315 [DOI] [PubMed] [Google Scholar]
- Cerutti H, Johnson AM, Gillham NW, Boynton JE (1997) Epigenetic silencing of a foreign gene in nuclear transformants of Chlamydomonas. Plant Cell 9: 925–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Hoff PL, Ferris P, Olson BJ, Miyagi A, Geng S, Umen JG (2013) Species and population level molecular profiling reveals cryptic recombination and emergent asymmetry in the dimorphic mating locus of C. reinhardtii. PLoS Genet 9: e1003724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43: 491–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan L, Nishii I, Harryman A, Buckley S, Howard A, Friedman NR, Miller SM (2007) The VARL gene family and the evolutionary origins of the master cell-type regulatory gene, regA, in Volvox carteri. J Mol Evol 65: 1–11 [DOI] [PubMed] [Google Scholar]
- Duncan L, Nishii I, Howard A, Kirk D, Miller SM (2006) Orthologs and paralogs of regA, a master cell-type regulatory gene in Volvox carteri. Curr Genet 50: 61–72 [DOI] [PubMed] [Google Scholar]
- Dyer TA. (1982) Methylation of chloroplast DNA in Chlamydomonas. Nature 298: 422–423 [DOI] [PubMed] [Google Scholar]
- Fang W, Si Y, Douglass S, Casero D, Merchant SS, Pellegrini M, Ladunga I, Liu P, Spalding MH (2012) Transcriptome-wide changes in Chlamydomonas reinhardtii gene expression regulated by carbon dioxide and the CO2-concentrating mechanism regulator CIA5/CCM1. Plant Cell 24: 1876–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, et al. (2010) Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci USA 107: 8689–8694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng TY, Chiang KS (1984) The persistence of maternal inheritance in Chlamydomonas despite hypomethylation of chloroplast DNA induced by inhibitors. Proc Natl Acad Sci USA 81: 3438–3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris PJ, Armbrust EV, Goodenough UW (2002) Genetic structure of the mating-type locus of Chlamydomonas reinhardtii. Genetics 160: 181–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris PJ, Goodenough UW (1987) Transcription of novel genes, including a gene linked to the mating-type locus, induced by Chlamydomonas fertilization. Mol Cell Biol 7: 2360–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris PJ, Waffenschmidt S, Umen JG, Lin H, Lee JH, Ishida K, Kubo T, Lau J, Goodenough UW (2005) Plus and minus sexual agglutinins from Chlamydomonas reinhardtii. Plant Cell 17: 597–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallaher SD, Fitz-Gibbon ST, Glaesener AG, Pellegrini M, Merchant SS (2015) Chlamydomonas genome resource for laboratory strains reveals a mosaic of sequence variation, identifies true strain histories, and enables strain-specific studies. Plant Cell 27: 2335–2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaesener AG, Merchant SS, Blaby-Haas CE (2013) Iron economy in Chlamydomonas reinhardtii. Front Plant Sci 4: 337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M. (1991) Transgenic expression of aminoglycoside adenine transferase in the chloroplast: a selectable marker of site-directed transformation of Chlamydomonas. Nucleic Acids Res 19: 4083–4089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Ballester D, Casero D, Cokus S, Pellegrini M, Merchant SS, Grossman AR (2010) RNA-seq analysis of sulfur-deprived Chlamydomonas cells reveals aspects of acclimation critical for cell survival. Plant Cell 22: 2058–2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough U, Lin H, Lee JH (2007) Sex determination in Chlamydomonas. Semin Cell Dev Biol 18: 350–361 [DOI] [PubMed] [Google Scholar]
- Grief C, O’Neill MA, Shaw PJ (1987) The zygote cell wall of Chlamydomonas reinhardtii: a structural, chemical and immunological approach. Planta 170: 433–445 [DOI] [PubMed] [Google Scholar]
- Grossman A. (2000) Acclimation of Chlamydomonas reinhardtii to its nutrient environment. Protist 151: 201–224 [DOI] [PubMed] [Google Scholar]
- Guo W, Fiziev P, Yan W, Cokus S, Sun X, Zhang MQ, Chen PY, Pellegrini M (2013) BS-Seeker2: a versatile aligning pipeline for bisulfite sequencing data. BMC Genomics 14: 774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris EH, Stern DB, Witman G, editors (2009) The Chlamydomonas Sourcebook. Elsevier/Academic Press, Amsterdam [Google Scholar]
- Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, et al. (2004) The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res 32: D258–D261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattman S, Kenny C, Berger L, Pratt K (1978) Comparative study of DNA methylation in three unicellular eucaryotes. J Bacteriol 135: 1156–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinnickel ML, Grossman AR (2013) The GreenCut: re-evaluation of physiological role of previously studied proteins and potential novel protein functions. Photosynth Res 116: 427–436 [DOI] [PubMed] [Google Scholar]
- Idoine AD, Boulouis A, Rupprecht J, Bock R (2014) The diurnal logic of the expression of the chloroplast genome in Chlamydomonas reinhardtii. PLoS One 9: e108760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S (2000) KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28: 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpowicz SJ, Prochnik SE, Grossman AR, Merchant SS (2011) The GreenCut2 resource, a phylogenomically derived inventory of proteins specific to the plant lineage. J Biol Chem 286: 21427–21439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk MM, Stark K, Miller SM, Müller W, Taillon BE, Gruber H, Schmitt R, Kirk DL (1999) regA, a Volvox gene that plays a central role in germ-soma differentiation, encodes a novel regulatory protein. Development 126: 639–647 [DOI] [PubMed] [Google Scholar]
- Kubo T, Abe J, Oyamada T, Ohnishi M, Fukuzawa H, Matsuda Y, Saito T (2008) Characterization of novel genes induced by sexual adhesion and gamete fusion and of their transcriptional regulation in Chlamydomonas reinhardtii. Plant Cell Physiol 49: 981–993 [DOI] [PubMed] [Google Scholar]
- Kuriyama H, Takano H, Suzuki L, Uchida H, Kawano S, Kuroiwa H, Kuroiwa T (1999) Characterization of Chlamydomonas reinhardtii zygote-specific cDNAs that encode novel proteins containing ankyrin repeats and WW domains. Plant Physiol 119: 873–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurvari V, Grishin NV, Snell WJ (1998) A gamete-specific, sex-limited homeodomain protein in Chlamydomonas. J Cell Biol 143: 1971–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck KF, Rostmann J, Grunow A (2002) Analysis of Chlamydomonas SF-assemblin by GFP tagging and expression of antisense constructs. J Cell Sci 115: 1511–1522 [DOI] [PubMed] [Google Scholar]
- Lee JH, Lin H, Joo S, Goodenough U (2008) Early sexual origins of homeoprotein heterodimerization and evolution of the plant KNOX/BELL family. Cell 133: 829–840 [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R (2010) Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2 6: 589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Misamore MJ, Snell WJ (2010) Membrane fusion triggers rapid degradation of two gamete-specific, fusion-essential proteins in a membrane block to polygamy in Chlamydomonas. Development 137: 1473–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez D, Casero D, Cokus SJ, Merchant SS, Pellegrini M (2011) Algal Functional Annotation Tool: a web-based analysis suite to functionally interpret large gene lists using integrated annotation and expression data. BMC Bioinformatics 12: 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv H, Qu G, Qi X, Lu L, Tian C, Ma Y (2013) Transcriptome analysis of Chlamydomonas reinhardtii during the process of lipid accumulation. Genomics 101: 229–237 [DOI] [PubMed] [Google Scholar]
- Matsuo M, Hachisu R, Tabata S, Fukuzawa H, Obokata J (2011) Transcriptome analysis of respiration-responsive genes in Chlamydomonas reinhardtii: mitochondrial retrograde signaling coordinates the genes for cell proliferation with energy-producing metabolism. Plant Cell Physiol 52: 333–343 [DOI] [PubMed] [Google Scholar]
- Matters GL, Goodenough UW (1992) A gene/pseudogene tandem duplication encodes a cysteine-rich protein expressed during zygote development in Chlamydomonas reinhardtii. Mol Gen Genet 232: 81–88 [DOI] [PubMed] [Google Scholar]
- Maul JE, Lilly JW, Cui L, dePamphilis CW, Miller W, Harris EH, Stern DB (2002) The Chlamydomonas reinhardtii plastid chromosome: islands of genes in a sea of repeats. Plant Cell 14: 2659–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant SS, Allen MD, Kropat J, Moseley JL, Long JC, Tottey S, Terauchi AM (2006) Between a rock and a hard place: trace element nutrition in Chlamydomonas. Biochim Biophys Acta 1763: 578–594 [DOI] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, et al. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R, Wu G, Deshpande RR, Vieler A, Gärtner K, Li X, Moellering ER, Zäuner S, Cornish AJ, Liu B, et al. (2010) Changes in transcript abundance in Chlamydomonas reinhardtii following nitrogen deprivation predict diversion of metabolism. Plant Physiol 154: 1737–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SM, Schmitt R, Kirk DL (1993) Jordan, an active Volvox transposable element similar to higher plant transposons. Plant Cell 5: 1125–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami SA, Goodenough UW (1978) Novel glycopolypeptide synthesis induced by gametic cell fusion in Chlamydomonas reinhardtii. J Cell Biol 77: 165–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S. (2010) Paternal inheritance of mitochondria in Chlamydomonas. J Plant Res 123: 163–170 [DOI] [PubMed] [Google Scholar]
- Nguyen AV, Thomas-Hall SR, Malnoë A, Timmins M, Mussgnug JH, Rupprecht J, Kruse O, Hankamer B, Schenk PM (2008) Transcriptome for photobiological hydrogen production induced by sulfur deprivation in the green alga Chlamydomonas reinhardtii. Eukaryot Cell 7: 1965–1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning J, Otto TD, Pfander C, Schwach F, Brochet M, Bushell E, Goulding D, Sanders M, Lefebvre PA, Pei J, et al. (2013) Comparative genomics in Chlamydomonas and Plasmodium identifies an ancient nuclear envelope protein family essential for sexual reproduction in protists, fungi, plants, and vertebrates. Genes Dev 27: 1198–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y. (2010) Uniparental inheritance of cpDNA and the genetic control of sexual differentiation in Chlamydomonas reinhardtii. J Plant Res 123: 149–162 [DOI] [PubMed] [Google Scholar]
- Nishimura Y, Misumi O, Kato K, Inada N, Higashiyama T, Momoyama Y, Kuroiwa T (2002) An mt(+) gamete-specific nuclease that targets mt(−) chloroplasts during sexual reproduction in C. reinhardtii. Genes Dev 16: 1116–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama R, Ito M, Yamaguchi Y, Koizumi N, Sano H (2002) A chloroplast-resident DNA methyltransferase is responsible for hypermethylation of chloroplast genes in Chlamydomonas maternal gametes. Proc Natl Acad Sci USA 99: 5925–5930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama R, Wada Y, Mibu M, Yamaguchi Y, Shimogawara K, Sano H (2004) Role of a nonselective de novo DNA methyltransferase in maternal inheritance of chloroplast genes in the green alga, Chlamydomonas reinhardtii. Genetics 168: 809–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchy N, Wu G, Newton L, Tsai CH, Chen J, Benning C, Farré EM, Shiu SH (2014) Prevalence, evolution, and cis-regulation of diel transcription in Chlamydomonas reinhardtii. G3 (Bethesda) 4: 2461–2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochnik SE, Umen J, Nedelcu AM, Hallmann A, Miller SM, Nishii I, Ferris P, Kuo A, Mitros T, Fritz-Laylin LK, et al. (2010) Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science 329: 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix JD. (2001) Assembly, function, and dynamics of the photosynthetic machinery in Chlamydomonas reinhardtii. Plant Physiol 127: 1394–1398 [PMC free article] [PubMed] [Google Scholar]
- Royer HD, Sager R (1979) Methylation of chloroplast DNAs in the life cycle of Chlamydomonas. Proc Natl Acad Sci USA 76: 5794–5798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano H, Grabowy C, Sager R (1981) Differential activity of DNA methyltransferase in the life cycle of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 78: 3118–3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmollinger S, Mühlhaus T, Boyle NR, Blaby IK, Casero D, Mettler T, Moseley JL, Kropat J, Sommer F, Strenkert D, et al. (2014) Nitrogen-sparing mechanisms in Chlamydomonas affect the transcriptome, the proteome, and photosynthetic metabolism. Plant Cell 26: 1410–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silflow CD, Lefebvre PA (2001) Assembly and motility of eukaryotic cilia and flagella: lessons from Chlamydomonas reinhardtii. Plant Physiol 127: 1500–1507 [PMC free article] [PubMed] [Google Scholar]
- Simon DF, Descombes P, Zerges W, Wilkinson KJ (2008) Global expression profiling of Chlamydomonas reinhardtii exposed to trace levels of free cadmium. Environ Toxicol Chem 27: 1668–1675 [DOI] [PubMed] [Google Scholar]
- Simon DF, Domingos RF, Hauser C, Hutchins CM, Zerges W, Wilkinson KJ (2013) Transcriptome sequencing (RNA-seq) analysis of the effects of metal nanoparticle exposure on the transcriptome of Chlamydomonas reinhardtii. Appl Environ Microbiol 79: 4774–4785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Meissner A (2013) DNA methylation: roles in mammalian development. Nat Rev Genet 14: 204–220 [DOI] [PubMed] [Google Scholar]
- Suzuki L, Woessner JP, Uchida H, Kuroiwa H, Yuasa Y, Waffenschmidt S, Goodenough UW, Kuroiwa T (2000) Zygote-specific protein with hydroxyproline-rich glycoprotein domains and lectin-like domains involved in the assembly of the cell wall of Chlamydomonas reinhardtii (Chlorophyta). J Phycol 36: 571–583 [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Vacquier VD (2002) The rapid evolution of reproductive proteins. Nat Rev Genet 3: 137–144 [DOI] [PubMed] [Google Scholar]
- Tam LW, Ranum PT, Lefebvre PA (2013) CDKL5 regulates flagellar length and localizes to the base of the flagella in Chlamydomonas. Mol Biol Cell 24: 588–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Blasing O, Gibon Y, Nagel A, Meyer S, Kruger P, Selbig J, Muller LA, Rhee SY, Stitt M (2004). MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP (2013) Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14: 178–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toepel J, Albaum SP, Arvidsson S, Goesmann A, la Russa M, Rogge K, Kruse O (2011) Construction and evaluation of a whole genome microarray of Chlamydomonas reinhardtii. BMC Genomics 12: 579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toepel J, Illmer-Kephalides M, Jaenicke S, Straube J, May P, Goesmann A, Kruse O (2013) New insights into Chlamydomonas reinhardtii hydrogen production processes by combined microarray/RNA-seq transcriptomics. Plant Biotechnol J 11: 717–733 [DOI] [PubMed] [Google Scholar]
- Uchida H, Kawano S, Sato N, Kuroiwa T (1993) Isolation and characterization of novel genes which are expressed during the very early stage of zygote formation in Chlamydomonas reinhardtii. Curr Genet 24: 296–300 [DOI] [PubMed] [Google Scholar]
- Uchida H, Suzuki L, Anai T, Doi K, Takano H, Yamashita H, Oka T, Kawano S, Tomizawa KI, Kawazu T, et al. (1999) A pair of invertedly repeated genes in Chlamydomonas reinhardtii encodes a zygote-specific protein whose expression is UV-sensitive. Curr Genet 36: 232–240 [DOI] [PubMed] [Google Scholar]
- Ueki N, Nishii I (2008) Idaten is a new cold-inducible transposon of Volvox carteri that can be used for tagging developmentally important genes. Genetics 180: 1343–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umen JG. (2011) Evolution of sex and mating loci: an expanded view from volvocine algae. Curr Opin Microbiol 14: 634–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umen JG, Goodenough UW (2001) Chloroplast DNA methylation and inheritance in Chlamydomonas. Genes Dev 15: 2585–2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woessner JP, Molendijk AJ, van Egmond P, Klis FM, Goodenough UW, Haring MA (1994) Domain conservation in several volvocalean cell wall proteins. Plant Mol Biol 26: 947–960 [DOI] [PubMed] [Google Scholar]
- Yang N, Xu RM (2013) Structure and function of the BAH domain in chromatin biology. Crit Rev Biochem Mol Biol 48: 211–221 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.