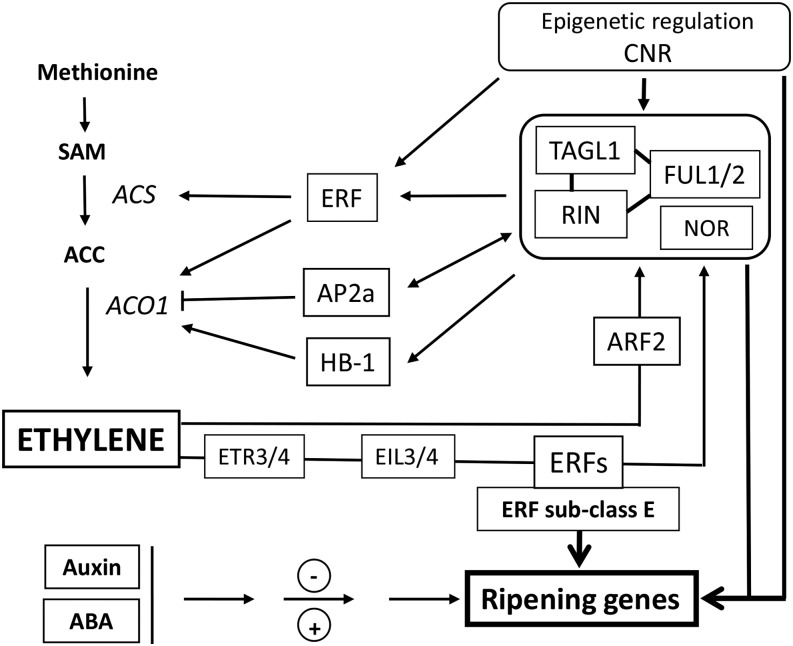
Figure 5.
Schematic overview of the multifactor regulatory network involved in ethylene biosynthesis and signaling during fruit development and ripening. RIN, TAGL1, and FUL1/2 are linked since they probably function as complexes of varying composition. The ripening master regulator NOR is placed in the same box. Along with CNR, these factors are master regulators of climacteric ripening. CNR affects the expression of RIN, LeHB1, SlAP2a, and SlTAGL1. FUL1 and FUL2 can potentially regulate ethylene biosynthesis, perception, and signaling genes. RIN promotes ripening via direct regulation of some transcription factors, such as ERFs. SlERF.B1 and SlERF.E1 are hypermethylated in cnr and rin mutants. ERFs regulate ethylene production in tomato by interaction with the promoters of ACO. Another transcription factor, LeHB-1, can bind in vivo to the promoter of ACO. The putative transcription factor Sl-AP2a was described as a negative regulator of fruit ripening and ethylene production. In addition, the control of ethylene biosynthesis can be regulated by RIN through direct interaction with the promoters of ACS2, ACS4, and ACO1. The ethylene biosynthesis pathway is controlled by a feedback mechanism, where ethylene regulates the expression of RIN. Moreover, there is evidence that ARFs also contribute to this complex feedback mechanism. ERF-type transcription factors are involved in fruit ripening through the control of ethylene and carotenoid biosynthesis pathways in tomato. Other hormones, such as Auxin and abscisic acid (ABA), also play a role in tuning fruit ripening. In particular, ARF2 was reported to be an essential component of the regulatory network controlling fruit ripening in tomato. Arrowheads represent positive regulatory interactions, and bar heads represent negative regulation. SAM, S-adenosyl-l-Met.