FIGURE 3:
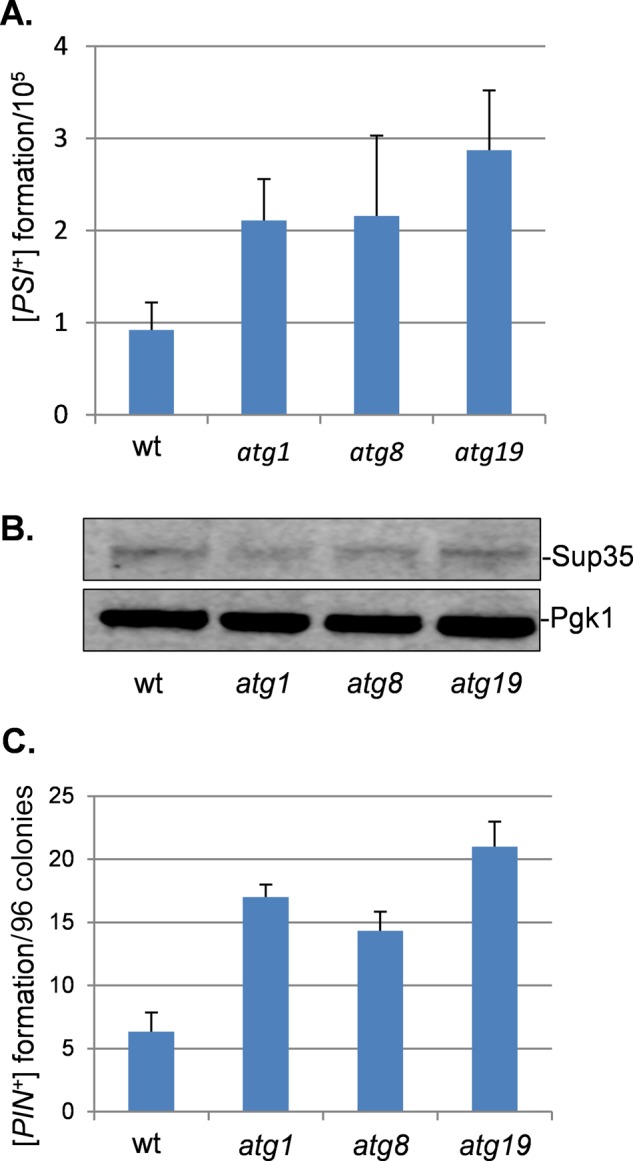
Increased frequency of de novo [PSI+] and [PIN+] prion formation in autophagy mutants. (A) [PSI+] prion formation was quantified in the wild-type and atg1, atg8, and atg19 mutant strains using an engineered ura3-14 allele, which contains the ade1-14 nonsense mutation inserted into the wild-type URA3 gene (Manogaran et al., 2006). [PSI+] prion formation was scored by growth on medium lacking uracil, indicative of decreased translational termination efficiency. [PSI+] formation was differentiated from nuclear gene mutations that give rise to uracil prototrophy by their irreversible elimination in GdnHCl. Data shown are the means of at least three independent biological repeat experiments expressed as the number of colonies per 105 viable cells. Error bars denote the SD. (B) Western blot analysis showing Sup35 protein levels in wild-type and atg1, atg8, and atg19 mutant strains. Blots were probed with α-Pgk1 as a loading control. (C) [PIN+] prion formation was quantified in wild-type and atg1, atg8, and atg19 mutant strains and is expressed as the number of [PIN+] colonies formed per 96 colonies examined. Data shown are the means of at least three independent biological experiments; error bars denote the SD.
