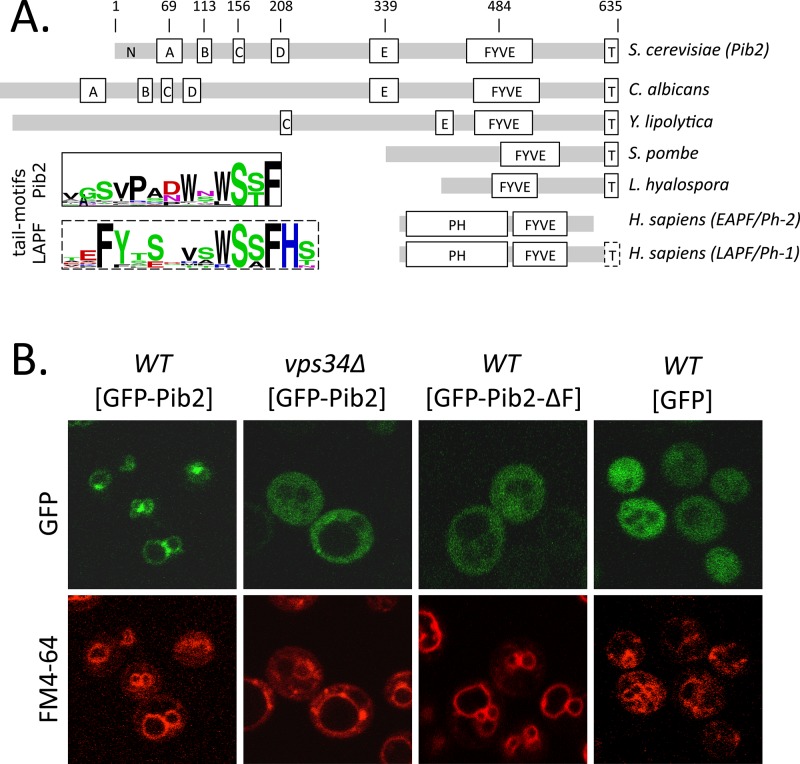
FIGURE 1:
Domain architecture and subcellular localization of Pib2. (A) Schematic diagrams of Pib2 from S. cerevisiae, other budding yeasts (C. albicans, Yarrowia lipolytica), fission yeast (Schizosaccharomyces pombe), a mushroom (Lichtheimia hyalospora), plus EAPF/phafin1 and LAPF/phafin2 (Homo sapiens), indicating the approximate positions of partially conserved motifs (A–E) and the universally conserved FYVE domain and tail motif (T). Gray bars indicate segments of poor conservation. Sequence logos of tail motifs from Pib2- and LAPF/phafin1 proteins are shown. (B) Fluorescence micrographs of wild-type and vps34∆ mutant strains of S. cerevisiae expressing GFP or GFP-Pib2 fusion protein (with and without the FYVE domain) after 30 min of staining with FM4-64 to reveal vacuolar and endosomal membranes.