Figure 1.
Structural Basis of SSB Recognition by PARP-1
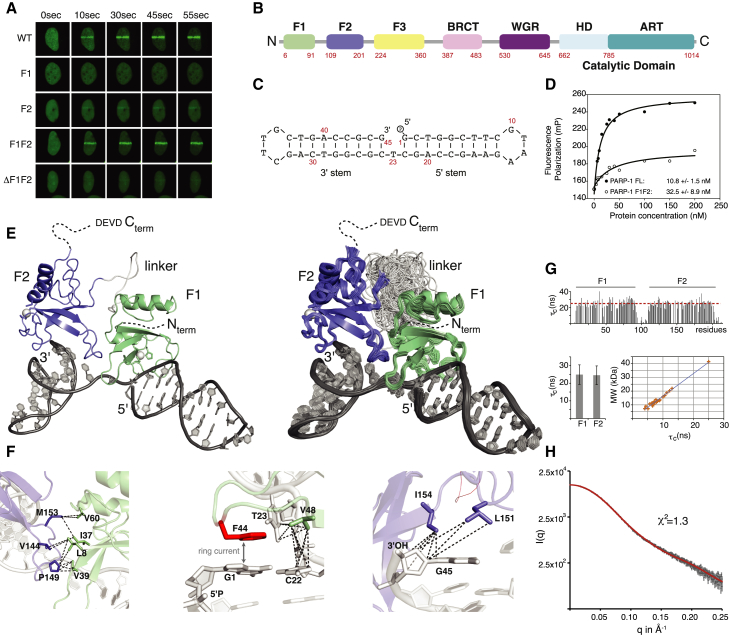
(A) Live-cell imaging shows recruitment of GFP-labeled PARP-1 and PARP-1 fragments to sites of laser-induced DNA damage. WT, wild-type.
(B) Domain structure of PARP-1.
(C) Gapped dumbbell DNA ligand used in this work as a mimic of an SSB.
(D) Fluorescence polarization experiments show that F1F2 binds the DNA ligand only about 3-fold less strongly than full-length PARP-1. FL, full-length.
(E) NMR/X-ray hybrid structure of F1F2 bound to an SSB. Overall views of (left) the lowest-energy structure, and (right) the ensemble of all 78 accepted structures (see Experimental Procedures and Supplemental Experimental Procedures for details of structure determination).
(F) Measured NOE contacts (dashed lines; Table S2) that define the F1-F2 interface (left), the hydrophobic interactions of F1 with the 5′ stem and T23 (center), and of F2 with the 3′ stem (right). The stacking interaction between F44 (red) and G1 was inferred from strong CSPs caused by their aromatic ring currents.
(G) Effective τc values obtained from 15N relaxation (TRACT) experiments with the F1F2 complex are consistent with a 40-kDa species (1:1 stoichiometry) in which both fingers bind simultaneously and the linker remains flexible.
(H) The experimental SAXS profile of the PARP-1 F1F2 dumbbell-DNA complex (3 mg.ml−1) agrees with the back-calculated SAXS profile averaged over the ensemble shown in (E).