Figure 2.
Interactions of F1 and F2 on Either Side of the SSB
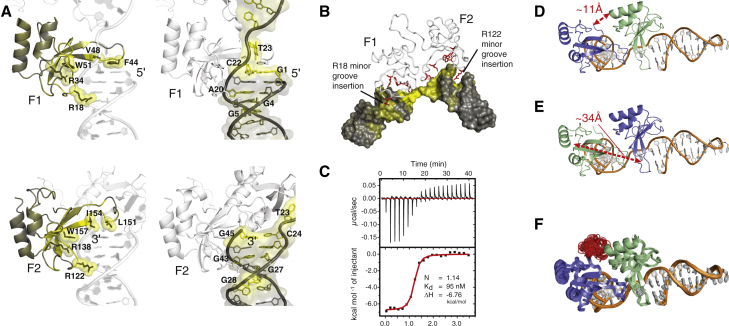
(A) CSPs on DNA binding mapped to the structure for protein amide groups (left) and DNA 1′-CH groups (right), for the F1-5′ DNA stem interface (top), and for the F2-3′ DNA stem interface (bottom). See Figure S2 for details and definitions of gray to yellow color ramps. Large CSPs occur for the key interacting protein residues labeled. For the DNA, CSPs match the 7-base pair footprints of the fingers on either side of the break (de Murcia and Ménissier de Murcia, 1994), with the largest effects at the exposed stem ends (C22-G1 and C24-G45) and sites of arginine insertions into the minor groove (Arg122 of F2 near C27, G28, and G43 in the 3′ stem and Arg18 of F1 near G4, G5, and A20 in the 5′ stem).
(B) CSPs measured for the DNA on F1F2 binding, illustrating the largest perturbations that occur at the DNA damage site. Protein side chains interacting with the DNA backbone are shown in red.
(C) Isothermal calorimetry shows that high-affinity binding of PARP-1 F1F2 to the gapped DNA dumbbell ligand occurs with 1:1 stoichiometry and is fully saturated at higher protein:DNA ratios (note that the apparent Kd is unreliable under these stoichiometric conditions).
(D and E) Schematics showing how reversing the directionality of the protein on the SSB would affect the F1-F2 linker (which was shown to be flexible by NMR; Figure 1G). If F1 bound the 3′ stem and F2 the 5′, then the distance spanned by the linker would increase from ∼11 Å in the actual complex (D) to ∼34 Å in the hypothetical reversed complex (E).
(F) Further calculations show that the observed arrangement of F1 and F2 is also consistent with the artificially shortened linker (Δ94–102). Ali et al. (2012) have reported that PARP-1 Δ94–102 localizes to laser-induced DNA damage in a very similar manner as wild-type protein in vivo.