Figure 3.
Structural Model of Full-Length PARP-1 Bound to a DNA Single-Strand Break
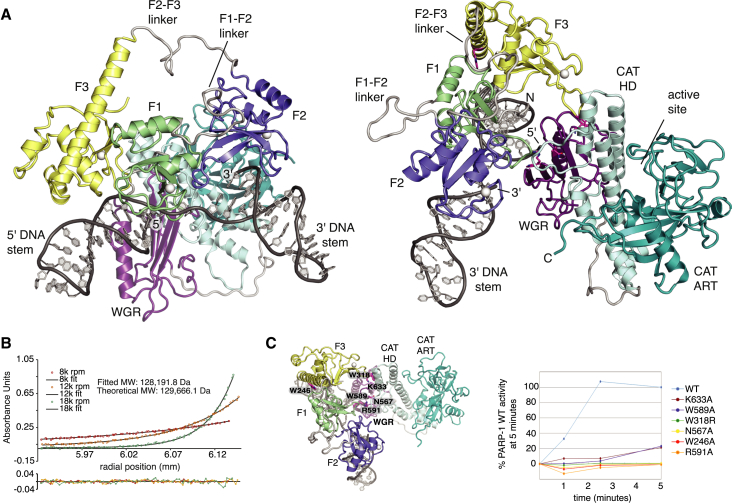
(A) Superposition of the hybrid structure of F1F2 bound to a DNA dumbbell and the previous crystal structure of F1, F3, and WGR-CAT bound to a DNA duplex (PDB: 4DQY) led directly to the domain arrangement shown (see also Figures S5g and S5h, Experimental Procedures, and Supplemental Experimental Procedures). The F1-F2 and F2-F3 linkers are flexible (see the NMR 15N relaxation data in Figure S5f), whereas the BRCT domain (not required for activity) and its linkers are omitted but may also adopt a wide variety of locations. The structure of PARP-1 on an SSB was corroborated using biophysical and mutational analysis (see B and C), NMR spectroscopy that elucidated its dynamic assembly process (Figure 4), and HXMS experiments (Dawicki-McKenna et al., 2015).
(B) Analytical ultracentrifugation shows that full-length PARP-1 binds to the gapped DNA dumbbell with a 1:1 stoichiometry. MW, molecular weight.
(C) Catalytic activities of wild-type PARP-1 and the designated mutants were assessed using a colorimetric activity assay (Supplemental Experimental Procedures) using the dumbbell gap DNA as the activating ligand.