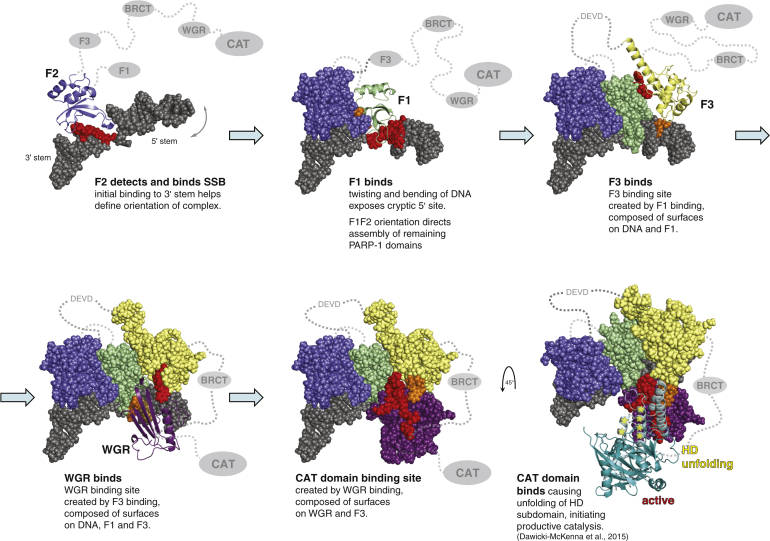
Figure 5.
Allosteric Activation Mechanism of PARP-1 by DNA Single-Strand Breaks
In the absence of DNA, PARP-1 domains behave independently, connected by disordered linkers. SSB recognition by F1 and F2 drives multidomain folding, which provides a cooperative switch for activation of the C-terminal catalytic domain (see main text); F3 and WGR are thereby positioned in the correct spatial orientation to trigger their ternary interaction with the catalytic domain. This relieves autoinhibition of the enzyme by causing local unfolding of the HD subdomain (indicated by stars in the figure), which is the subject of the accompanying paper by Dawicki-McKenna (2015). Interfacial residues on different components are colored red and orange. DEVD indicates the caspase cleavage site between F2 and F3. In this figure, we show the pathway as a sequence of discrete steps, but, in reality, they are probably not fully separated. For instance, although F2 likely initiates binding, F1 also co-operates in high-affinity DNA damage recognition. Similarly, although we show F3 binding ahead of WGR, in principle, these events could occur in either order or both. Nevertheless, our data show that elements of the pathway represent intermediate steps because they can occur in isolation. For instance, F1F2 binds an SSB and achieves directional selectivity on its own, and interactions of F3 and WGR occur with the F1F2-DNA complex in the absence of the CAT domain. BRCT, BRCA-1 C terminus.