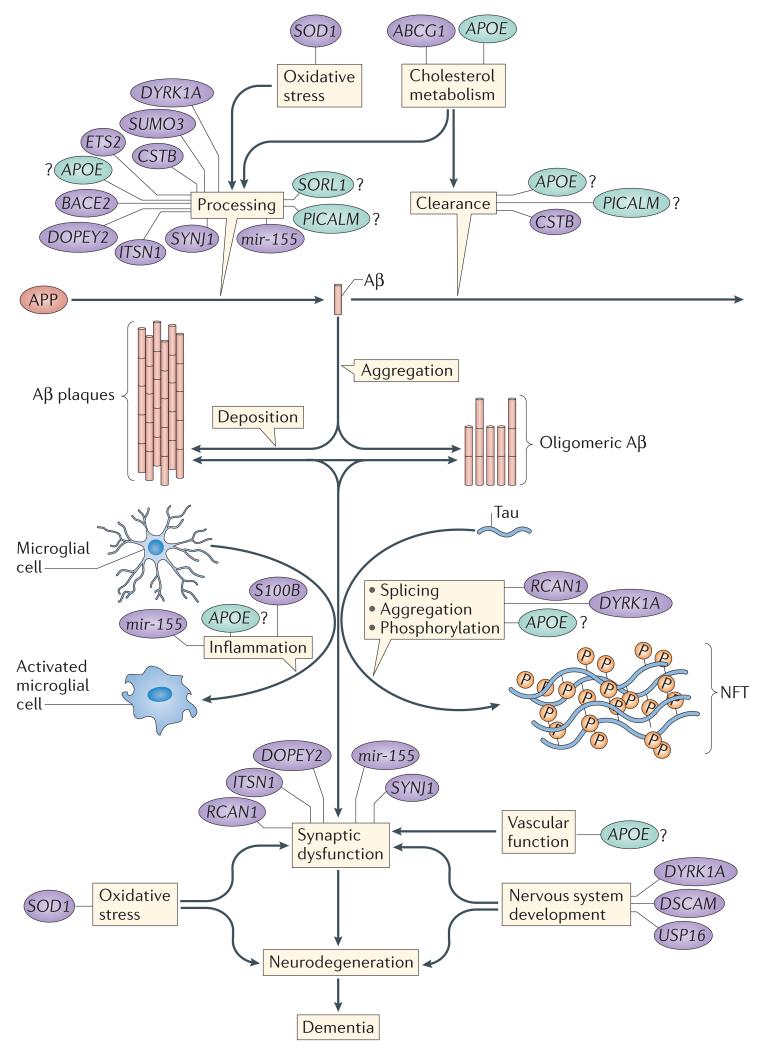
Figure 3. Schematic of suggested mechanisms that are important in AD-DS and their related genes.
Several genes may modulate processes that are relevant to the development of Alzheimer disease in people with Down syndrome (AD-DS); these include non-chromosome 21 genes, such as apolipoprotein E (APOE; which could alter disease by influencing cholesterol metabolism and possibly many other pathways), phosphatidylinositol-binding clathrin assembly protein (PICALM), sortilin-related receptor 1 (SORL1; which may influence disease via the endocytosis system and amyloid precursor protein (APP) processing) and microtubule-associated protein tau (MAPT). Tau aggregates to form neurofibrillary tangles (NFTs). Numerous chromosome 21 genes have also been suggested to influence the development of AD-DS, including genes that may influence APP processing and synaptic function via their role in the secretory–endosome system (including cystatin B (CSTB), DOPEY2, synaptojanin 1 (SYNJ1), intersectin 1 (ITSN1) and the microRNA gene mir-155), APP processing (including small ubiquitin-like modifier 3 (SUMO3), ETS2 and beta-site APP-cleaving enzyme 2 (BACE2)), cholesterol metabolism (including ATP-binding cassette G1 (ABCG1)), cellular signalling and tau phosphorylation (including dual-specificity tyrosine-phosphorylation-regulated kinase 1A (DYRK1A) and regulator of calcineurin 1 (RCAN1)), inflammation (including mir-155 and S100 calcium-binding protein beta (S100B)), synaptic function (including DOPEY2, SYNJ1, ITSN1, RCAN1 and mir-155), neurodevelopment (including ubiquitin-specific peptidase 16 (USP16), DYRK1A and DS cell adhesion molecule (DSCAM)) and oxidative stress (superoxide dismutase 1 (SOD1)). The relative importance of these processes to the development of dementia in AD-DS remains unclear and constitutes an area for future study. Chromosome 21 genes and gene products are shown in purple; non-chromosome 21 genes and gene products are shown in green. Aβ, amyloid-β.