Fig. 1.
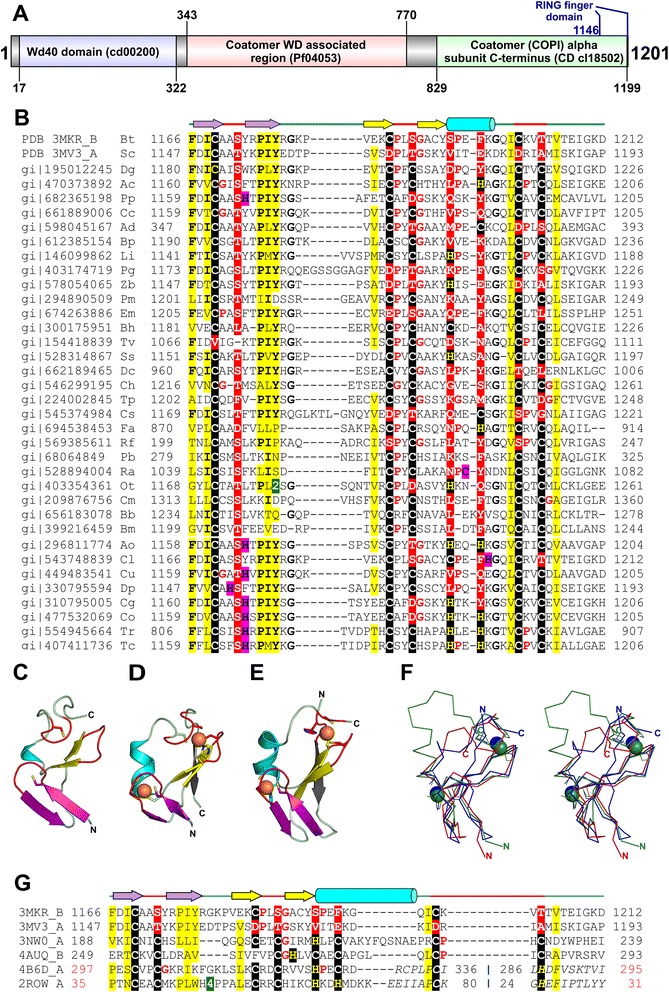
Structure and sequence comparison of α-COP and other RING fingers. a Domain diagram of S. cerevisiae α-COP. Domain boundaries were obtained by referring to CDD (Additional file 1). Position of the manually delineated RING finger domain is indicated in blue. b Structure-based multiple sequence alignment of RING domain from α-COP homologs. c RING domain of α-COP (PDBid 3MKR_B) (d) A classical RING finger domain from Baculoviral IAP repeat-containing protein 7 (PDBid 4AUQ_B) (e) A circularly permuted RING finger from Rac GTPase-activating protein 1 (PDBid 4B6D_A). The RING fingers are colored alike, zinc-binding knuckles: red, primary β-hairpin: yellow, α-helix: cyan, zinc knuckle containing β-hairpin: violet, the additional β-strand: gray, zinc ion: orange sphere. The N- and C-termini are labeled ‘N’ and ‘C’, respectively. Side chains of metal-chelating residues are shown in stick representation. f Stereo diagram of the superimposition of the α-COP RING finger (PDBid 3MV3_A; colored red), RING finger domain of Non-SMC element 1 homolog (PDBid 3NW0_B; colored green) and C1 domain of Rac GTPase-activating protein 1 (PDBid 4B6D_A; colored blue). The Cα backbone trace is shown for each structure, and bound zinc ions are shown as spheres in the respective colors. g Structure-based sequence alignment of RING domain of α-COP (PDBid 3MKR_B, 3MV3_A), RING finger domains (PDBid 3NW0_A, 4AUQ_B) and circularly permuted RING fingers (PDBid 4B6D_A, 2ROW_A). For panels (b, g) PDBid/Gene identification (gi) number, the first and the last residue numbers of the sequences depicted in the alignment are indicated for each sequence. The secondary structure elements of the RING finger domain are indicated above the alignment. The potential metal-binding ligands are boxed in black, non-metal-binding residues at the same position are boxed in red and adjacent residues that could potentially chelate metal ion are boxed in magenta. Conserved residues are in bold. Small aminoacids (Gly, Pro) in the vicinity of the zinc-chelating ligands are colored red. Uncharged residues (all aminoacids except Asp, Glu, Lys and Arg) in mostly hydrophobic sites are highlighted yellow. Some insertions are not shown and the number of omitted residues is specified by numbers boxed in green. The sequence numbers in the region of permutation in panel (g) are shown in red and regions of circular permutation are separated by a ‘|’ symbol. In panel (b) the PDBid/gi number is followed by organism name abbreviation as follows: Bt- Bos taurus, Sc- Saccharomyces cerevisiae, Dg- Drosophila grimshawi, Ac- Acanthamoeba castellanii str. Neff, Pp- Pseudogymnoascus pannorum VKM F-4516 (FW-969), Cc- Coffea canephora, Ad- Auricularia delicata TFB-10046 SS5, Bp- Bathycoccus prasinos, Li- Leishmania infantum JPCM5, Pg- Puccinia graminis f. sp. tritici CRL 75-36-700-3, Zb- Zygosaccharomyces bailii ISA1307, Pm- Perkinsus marinus ATCC 50983, Em- Echinococcus multilocularis, Bh- Blastocystis hominis, Tv- Trichomonas vaginalis G3, Ss- Schizosaccharomyces cryophilus OY26, Dc- Diaphorina citri, Ch- Chondrus crispus, Tp- Thalassiosira pseudonana CCMP1335, Cs- Coccomyxa subellipsoidea C-169, Fa- Fonticula alba, Rf- Reticulomyxa filose, Pb- Plasmodium berghei ANKA, Ra- Rozella allomycis CSF55, Ot- Oxytricha trifallax, Cm- Cryptosporidium muris RN66, Bb- Babesia bigemina, Bm- Babesia microti strain RI, Ao- Arthroderma otae CBS 113480, Cl- Columba livia, Cu- Cucumis sativus, Dp- Dictyostelium purpureum, Cg- Colletotrichum graminicola M1.001, Co- Colletotrichum orbiculare MAFF 240422, Tr- Trypanosoma rangeli SC58, Tc- Trypanosoma cruzi marinkellei
