Figure 3.
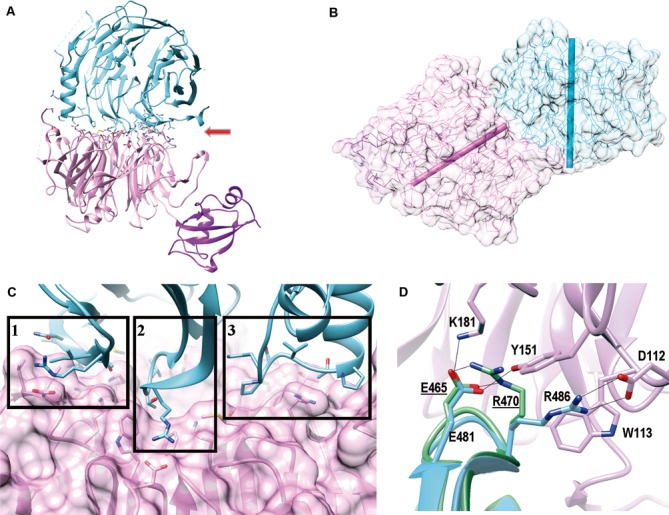
The β-propellers of ChErb1 and ChYtm1 interact in a novel manner. (A) Ribbon representation of the dimer shows that ChYtm1 (pink) binds to the C-terminal domain of ChErb1 (blue) through the top surface of the β-propeller (red arrow). UBL domain of ChYtm1 (purple) does not participate in the interaction. Side chains of interacting residues are shown. (B) The central axes of both β-propellers (shown as bars, ChYtm1:pink; ChErb1: blue) form an angle of 55°. (C) Three areas of ChErb1432–801 (blue) contact the β-propeller of ChYtm1 (pink): (i) Strand 1d from the blade 7 establishes non-conserved interactions with the knob that appears between blades 6 and 7 of ChYtm1. (ii) A well conserved loop ‘c–d’ from blade 1 of ChErb1 binds to the central channel of ChYtm1. (iii) The insertion from blade 2 of ChErb1 mediates binding to an extended loop from ChYtm1 on one side of the propeller. (D) Superimposition of the β-propeller of Erb1 alone (PDB: 4U7A, in green) shows that upon binding to ChYtm1 (pink) the interacting residues from ChErb1 (blue) reorganize and force R486 of ChErb1 toward salt-bridge formation with D112 of ChYtm1. Electrostatic interactions are shown as black lines. Labels corresponding to the residues of Erb1 from yeast are underlined.
