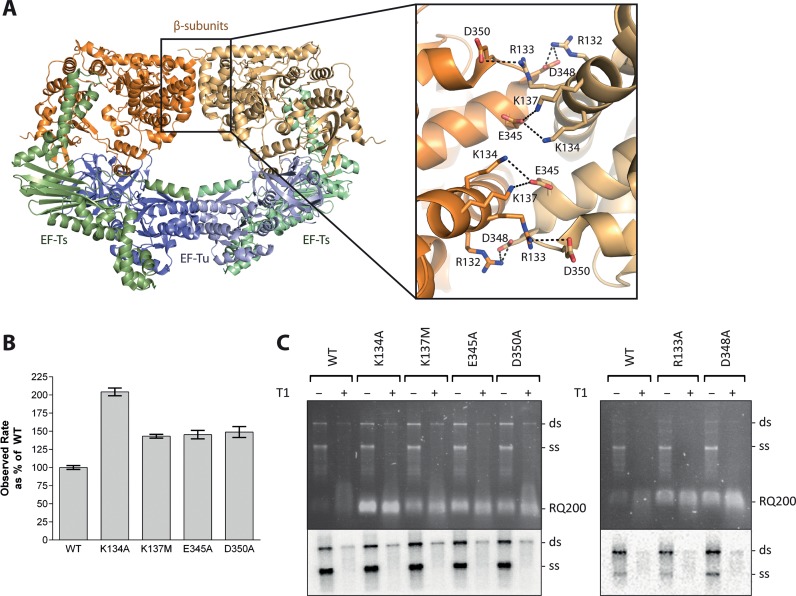
Figure 1.
Mutational analysis of residues at the dimerization interface of the Qβ replicase core complex involved in salt-bridge formation. (A) Structure of the dimer of the Qβ replicase core complex consisting of the viral β-subunit (orange) and the host proteins EF-Tu (blue) and EF-Ts (green). The two halves of the dimer are shown in different shades of these colours. The dimerization interface between two β-subunits is highlighted in the inset figure illustrating residues involved in salt bridges. (B) Results of an in vitro replication assay using the RQ139 template. Product formation was followed measuring SYBR green II fluorescence, and the reaction rates were deduced from the exponential phases of the real-time reaction curves and related to the activity of the wild-type Qβ replicase complex (mean set to 100%). Error bars indicate standard deviations for n = 3. Controls included reaction mixtures without Qβ replicase or RNA and reaction mixtures without rNTP to allow quantification of background fluorescence, which was subtracted from the measurements on complete reactions. (C) Results from an in vitro replication assay using the Qβ (+)-RNA template. The reaction was performed in low salt and lasted for 10 min before quenching. Replication products were either digested with T1 ribonuclease prior to phenol/chloroform extraction (+) or not (−). The genomic replication products were separated by agarose gel electrophoresis. The gel was stained with ethidium bromide and photographed under UV light (top). [32P]-labeled product bands were visualized after exposure to a storage phosphor screen (bottom). The positions of single- (ss) and double-stranded (ds) genomic Qβ RNA, as well as a contaminating, replicative RNA (RQ200) are indicated. The lower part of the gel containing RQ200 RNA was removed before exposure to the storage phosphor screen to avoid interference with the signal from the labeled genomic RNA. The assay was repeated three times, and a representative result is shown.