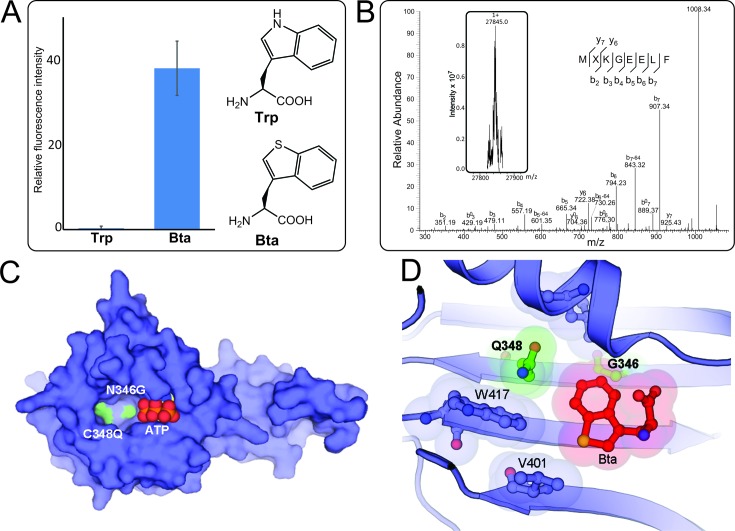
Figure 1.
The BtaRS evolved from PylRS bearing Asn346Gly and Cys348Gln mutations discriminates Bta from Trp. (A) Suppression of the UAG codon at position 2 of sfGFP-2UAG mRNA supported by BtaRS in the presence of Bta or Trp was measured by fluorescence intensity. (B) Molecular weight determination of the protein sfGFP-Bta and its MXKGEELF (X denotes Bta) fragment. The deconvoluted singly charged ESI-MS spectrum of sfGFP-Bta by FT-ICR MS (inserted Fig. and see Supplementary Figure S4 for the full MS image) and tandem mass spectra of the MXKGEELF (X denotes Bta) fragment from full-length sfGFP-Bta protein are shown. The full-length sfGFP-Bta protein was expressed using the BtaRS•tRNAPylCUA pair in the presence of 1 mM Bta. Calculated molecular weight of full-length sfGFP-Bta protein is 27 845 Da ([M+H]1+); we found 27 845 Da. b0 or y0 stands for b-H2O or y-H2O. (C) The overall structure of BtaRS in the absence of the Bta shows the positions of Asn346Gly and Cys348Gln mutations in the active site pocket. The side chains of Asn346Gly/Cys348Gln double mutations are shown in green, and the bound ATP is in combination of red and orange. (D) The anatomy of the BtaRS active site illustrates the substrate recognition. Mutations from the parent PylRS are indicated in bold, and the mutated side chains are shown in green. The shadows indicate the van der Waals radius of Bta and the surrounding residues that are involved in substrate recognition.