Figure 1.
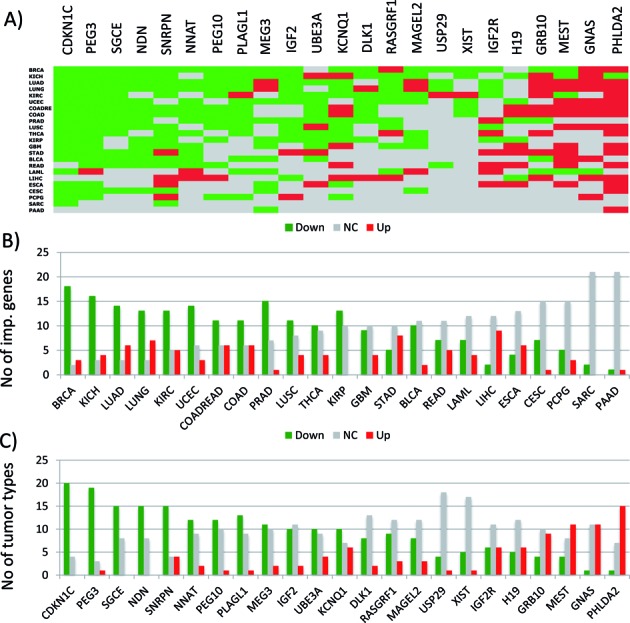
Expression level changes of imprinted genes in human cancers. (A) Expression levels of imprinted genes (represented by columns) were compared between the primary tumor and normal solid tissue cells of various cancers (represented by rows) using TCGA database (The Cancer Genome Atlas). Statistically significant down- and upregulations were indicated by green and red boxes, respectively, while no significant change with gray boxes. XIST has been included in this series of analyses due to its 50% DNA methylation levels in females although it is not an imprinted gene. The actual table used for this image is available as Supplementary Data 1. (B) This graph summarizes how many imprinted genes show down and upregulations for each cancer type. (C) This graph summarizes in how many cancer types the expression levels of each imprinted gene are changed between the tumor and normal cells. Individual cancer types are represented with the following abbreviations: BRCA (breast invasive carcinoma), KICH (kidney chromophobe), LUAD (lung adenocarcinoma), LUNG (lung cancer), KIRC (kidney renal clear cell carcinoma), UCEC (uterine corpus endometrioid carcinoma), COADREAD (colon and rectum adenocarcinoma), COAD (colon adenocarcinoma), PRAD (prostate adenocarcinoma), LUSC (lung squamous cell carcinoma), THCA (thyroid carcinoma), KIRP (kidney renal papillary cell carcinoma), GBM (glioblastoma multiforme), STAD (stomach adenocarcinoma), BLCA (bladder urothelial carcinoma), READ (rectum adenocarcinoma), LAML (acute myeloid leukemia), LIHC (liver hepatocellular carcinoma), ESCA (esophageal carcinoma), CESC (cervical squamous cell carcinoma and endocervical adenocarcinoma), PCPG (pheochromocytoma and paraganglioma), SARC (sarcoma) and PAAD (pancreatic adenocarcinoma).
